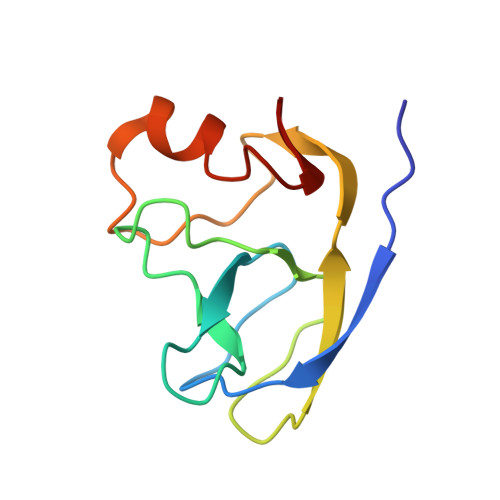
Crystal structure of a truncated version of the phage lambda protein gpD.
Chang, C., Plueckthun, A., Wlodawer, A.(2004) Proteins 57: 866-868
- PubMed: 15317025
- DOI: https://doi.org/10.1002/prot.20254
- Primary Citation of Related Structures:
1TCZ - Macromolecular Crystallography Laboratory, National Cancer Institute at Frederick, Frederick, Maryland 21702, USA.
Organizational Affiliation: