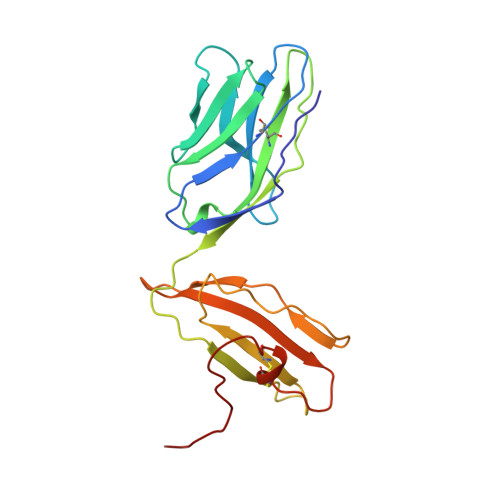
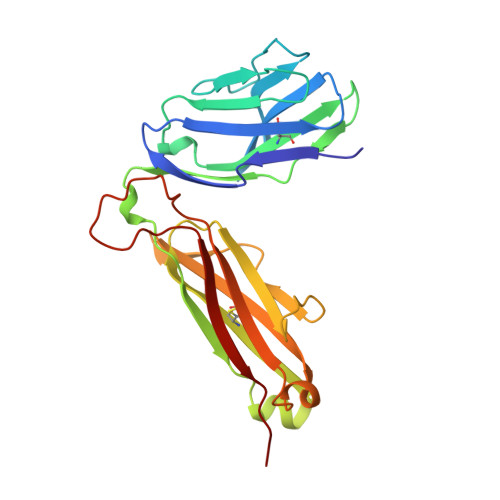
An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex.
Garcia, K.C., Degano, M., Stanfield, R.L., Brunmark, A., Jackson, M.R., Peterson, P.A., Teyton, L., Wilson, I.A.(1996) Science 274: 209-219
- PubMed: 8824178
- DOI: https://doi.org/10.1126/science.274.5285.209
- Primary Citation of Related Structures:
1TCR - PubMed Abstract:
The central event in the cellular immune response to invading microorganisms is the specific recognition of foreign peptides bound to major histocompatibility complex (MHC) molecules by the alphabeta T cell receptor (TCR). The x-ray structure of the complete extracellular fragment of a glycosylated alphabeta TCR was determined at 2.5 angstroms, and its orientation bound to a class I MHC-peptide (pMHC) complex was elucidated from crystals of the TCR-pMHC complex. The TCR resembles an antibody in the variable Valpha and Vbeta domains but deviates in the constant Calpha domain and in the interdomain pairing of Calpha with Cbeta. Four of seven possible asparagine-linked glycosylation sites have ordered carbohydrate moieties, one of which lies in the Calpha-Cbeta interface. The TCR combining site is relatively flat except for a deep hydrophobic cavity between the hypervariable CDR3s (complementarity-determining regions) of the alpha and beta chains. The 2C TCR covers the class I MHC H-2Kb binding groove so that the Valpha CDRs 1 and 2 are positioned over the amino-terminal region of the bound dEV8 peptide, the Vbeta chain CDRs 1 and 2 are over the carboxyl-terminal region of the peptide, and the Valpha and Vbeta CDR3s straddle the peptide between the helices around the central position of the peptide.
- Department of Molecular Biology and the Skaggs Institute of Chemical Biology, Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: