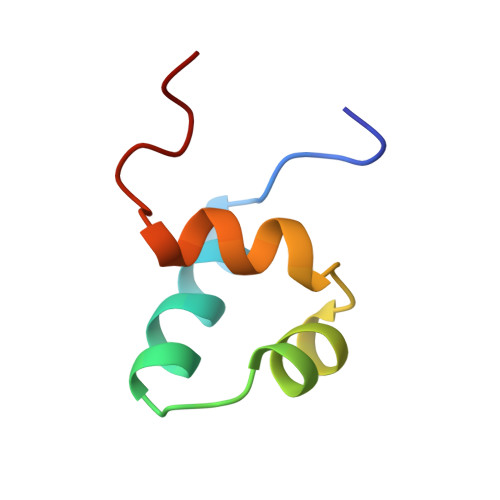
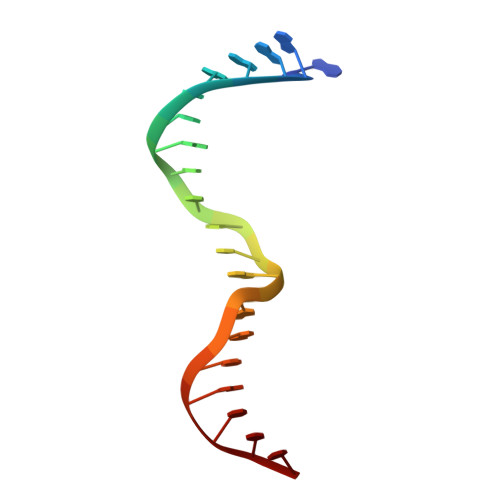
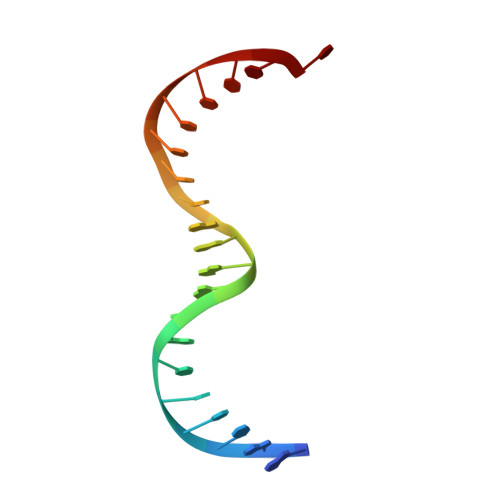
Crystal structure of the specific DNA-binding domain of Tc3 transposase of C.elegans in complex with transposon DNA.
van Pouderoyen, G., Ketting, R.F., Perrakis, A., Plasterk, R.H., Sixma, T.K.(1997) EMBO J 16: 6044-6054
- PubMed: 9312061
- DOI: https://doi.org/10.1093/emboj/16.19.6044
- Primary Citation of Related Structures:
1TC3 - PubMed Abstract:
The crystal structure of the complex between the N-terminal DNA-binding domain of Tc3 transposase and an oligomer of transposon DNA has been determined. The specific DNA-binding domain contains three alpha-helices, of which two form a helix-turn-helix (HTH) motif. The recognition of transposon DNA by the transposase is mediated through base-specific contacts and complementarity between protein and sequence-dependent deformations of the DNA. The HTH motif makes four base-specific contacts with the major groove, and the N-terminus makes three base-specific contacts with the minor groove. The DNA oligomer adopts a non-linear B-DNA conformation, made possible by a stretch of seven G:C base pairs at one end and a TATA sequence towards the other end. Extensive contacts (seven salt bridges and 16 hydrogen bonds) of the protein with the DNA backbone allow the protein to probe and recognize the sequence-dependent DNA deformation. The DNA-binding domain forms a dimer in the crystals. Each monomer binds a separate transposon end, implying that the dimer plays a role in synapsis, necessary for the simultaneous cleavage of both transposon termini.
- Division of Molecular Carcinogenesis, Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.
Organizational Affiliation: