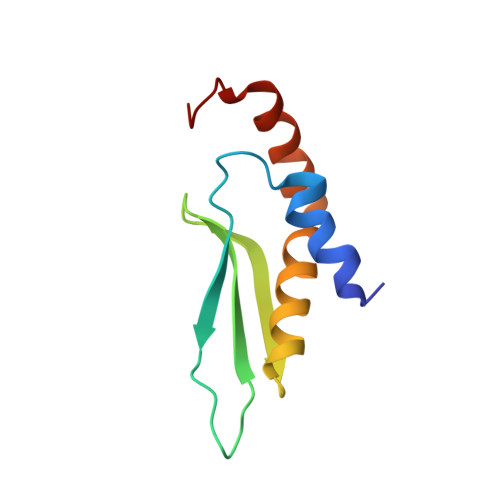
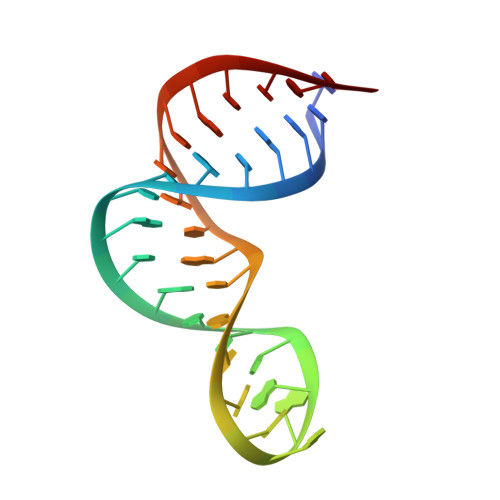
Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III.
Wu, H., Henras, A., Chanfreau, G., Feigon, J.(2004) Proc Natl Acad Sci U S A 101: 8307-8312
- PubMed: 15150409
- DOI: https://doi.org/10.1073/pnas.0402627101
- Primary Citation of Related Structures:
1T4L - PubMed Abstract:
Specific recognition of double-stranded RNA (dsRNA) by dsRNA-binding domains (dsRBDs) is involved in a large number of biological and regulatory processes. Although structures of dsRBDs in complex with dsRNA have revealed how they can bind to dsRNA in general, these do not explain how a dsRBD can recognize specific RNAs. Rnt1p, a member of the RNase III family of dsRNA endonucleases, is a key component of the Saccharomyces cerevisiae RNA-processing machinery. The Rnt1p dsRBD has been implicated in targeting this endonuclease to its RNA substrates, by recognizing hairpins closed by AGNN tetraloops. We report the solution structure of Rnt1p dsRBD complexed to the 5' terminal hairpin of one of its small nucleolar RNA substrates, the snR47 precursor. The conserved AGNN tetraloop fold is retained in the protein-RNA complex. The dsRBD contacts the RNA at successive minor, major, and tetraloop minor grooves on one face of the helix. Surprisingly, neither the universally conserved G nor the highly conserved A are recognized by specific hydrogen bonds to the bases. Rather, the N-terminal helix fits snugly into the minor groove of the RNA tetraloop and top of the stem, interacting in a non-sequence-specific manner with the sugar-phosphate backbone and the two nonconserved tetraloop bases. Mutational analysis of residues that contact the tetraloop region show that they are functionally important for RNA processing in the context of the entire protein in vivo. These results show how a single dsRBD can convey specificity for particular RNA targets, by structure specific recognition of a conserved tetraloop fold.
- Department of Chemistry and Biochemistry and Molecular Biology Institute, University of California, Los Angeles, CA 90095-1569, USA.
Organizational Affiliation: