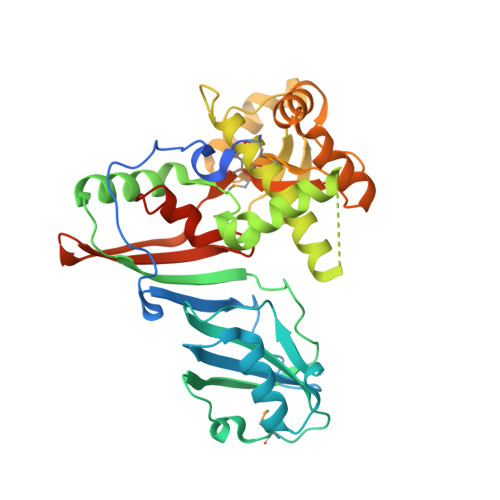
X-ray structure of tRNA pseudouridine synthase TruD reveals an inserted domain with a novel fold
Ericsson, U.B., Nordlund, P., Hallberg, B.M.(2004) FEBS Lett 565: 59-64
- PubMed: 15135053
- DOI: https://doi.org/10.1016/j.febslet.2004.03.085
- Primary Citation of Related Structures:
1SZW - PubMed Abstract:
Pseudouridine synthases catalyse the isomerisation of uridine to pseudouridine in structural RNA. The pseudouridine synthase TruD, that modifies U13 in tRNA, belongs to a recently identified and large family of pseudouridine synthases present in all kingdoms of life. We report here the crystal structure of Escherichia coli TruD at 2.0 A resolution. The structure reveals an overall V-shaped molecule with an RNA-binding cleft formed between two domains: a catalytic domain and an insertion domain. The catalytic domain has a fold similar to that of the catalytic domains of previously characterised pseudouridine synthases, whereas the insertion domain displays a novel fold.
- Department of Biochemistry and Biophysics, Stockholm University, Roslagstullsbacken 15, SE-114 21 Stockholm, Sweden.
Organizational Affiliation: