Determination of the three-dimensional solution structure of noxiustoxin: analysis of structural differences with related short-chain scorpion toxins.
Dauplais, M., Gilquin, B., Possani, L.D., Gurrola-Briones, G., Roumestand, C., Menez, A.(1995) Biochemistry 34: 16563-16573
- PubMed: 8527429
- DOI: https://doi.org/10.1021/bi00051a004
- Primary Citation of Related Structures:
1SXM - PubMed Abstract:
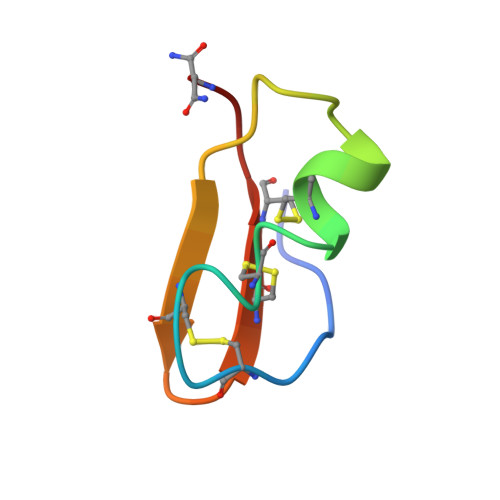
The 3D structure of noxiustoxin, the first identified scorpion toxin acting on K+ channels, has been elucidated by NMR and molecular modeling. Thirty-nine solution structures were calculated using 572 distance and 42 dihedral restraints. The average atomic rms deviation between the refined structures and the mean structure is 0.75 A for the backbone atoms. Noxiustoxin adopts a alpha/beta scaffold constituted of a three-stranded beta-sheet (residues 2-3, 25-30, 33-38) linked to a helix (residues 10-20) through two disulfide bridges. A comparison between the 3D structure of noxiustoxin and those of other structurally and functionally related scorpion toxins (charybdotoxin, PO5-NH2, kaliotoxin) revealed a bending capacity of the helix and a variability in the relative orientations between the helix and the beta-sheet. These two features highlight the plasticity of the alpha/beta scaffold and offer a structural explanation for the capacity of the fold to accommodate an additional alanine residue in the Gly-x-Cys pattern of a previously proposed consensus sequence [Bontems et al. (1991) Science 254, 1521-1523]. Our structural data also emphasize the possibility that the beta-sheet of NTX is implicated in the capacity of NTX to recognize voltage-dependent K+ channels.
- Département d'Ingénierie et d'Etude des Protéines, CE-Saclay, Gif-sur-Yvette, France.
Organizational Affiliation: