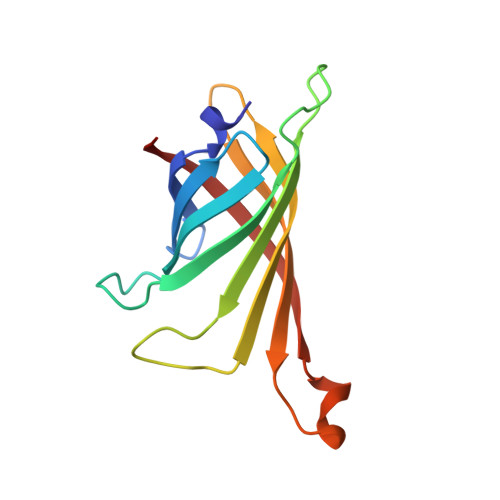
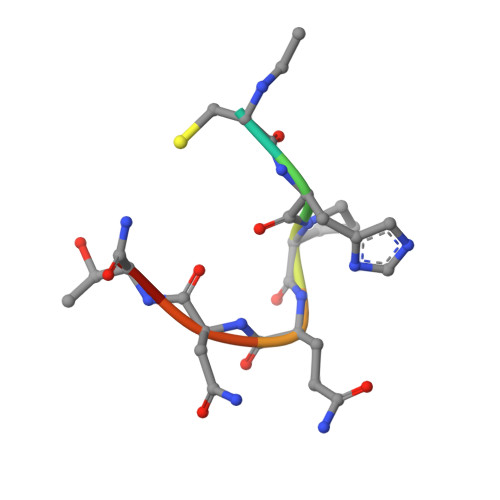
Topochemical catalysis achieved by structure-based ligand design.
Katz, B.A., Cass, R.T., Liu, B., Arze, R., Collins, N.(1995) J Biological Chem 270: 31210-31218
- PubMed: 8537386
- DOI: https://doi.org/10.1074/jbc.270.52.31210
- Primary Citation of Related Structures:
1STR, 1STS - PubMed Abstract:
Recently, a cyclic peptide ligand, cyclo-Ac-[CHPQG-PPC]-NH2, that binds to streptavidin with high affinity was discovered by screening phage libraries. From the streptavidin-bound crystal structures of cyclo-Ac-[CHPQGPPC]-NH2 and of a related but more weakly binding linear ligand, FSHPQNT, we designed linear thiol-containing streptavidin binding ligands, FCH-PQNT-NH2 and Ac-CHPQNT-NH2, which are dimerized catalytically by the streptavidin crystal lattice of space group I222, as demonstrated by high performance liquid chromatography and mass spectrometry. The catalytic dimerization relies on presentation of the ligand thiols toward one another in the lattice. The streptavidin crystal lattice-mediated catalysis achieved by structure-based design is the first example of catalysis of a chemical reaction by a protein crystal lattice. The spontaneous and crystal catalyzed rates of disulfide formation were determined by high performance liquid chromatography at pH 3.1, 4.0, 5.0, and 6.0. The ratio of the catalyzed to uncatalyzed rate was maximal at pH 3.1 (kcat/kuncat = 3.8), diminishing to 1.2 at pH 6.0. The crystal structures of the streptavidin-bound dimerized peptide ligands, FCHPQNT-NH2 dimer at 1.95 A and Ac-CHPQNT-NH2 dimer at 1.80 A, are described and compared with the structures of streptavidin-bound FSHPQNT monomer and cyclo-Ac-[CHPQGPPC]-NH2 dimer.
- Arris Pharmaceutical Corporation, South San Francisco, California 94080, USA.
Organizational Affiliation: