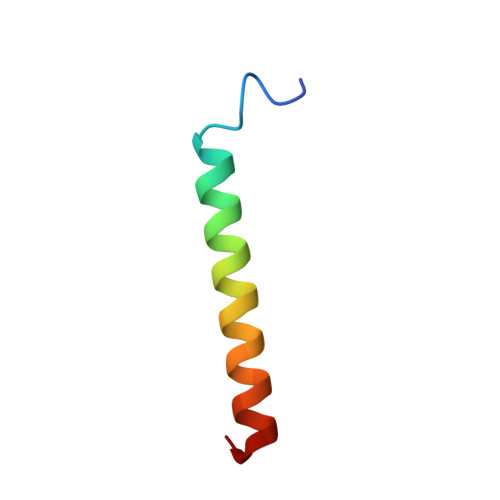
The NMR structure of the pulmonary surfactant-associated polypeptide SP-C in an apolar solvent contains a valyl-rich alpha-helix.
Johansson, J., Szyperski, T., Curstedt, T., Wuthrich, K.(1994) Biochemistry 33: 6015-6023
- PubMed: 8180229
- DOI: https://doi.org/10.1021/bi00185a042
- Primary Citation of Related Structures:
1SPF - PubMed Abstract:
The nuclear magnetic resonance (NMR) structure of the pulmonary surfactant-associated lipoplypeptide C (SP-C) was determined in a mixed solvent of C2H3Cl/C2H3OH/ 1 M HCl 32:64:5 (v/v). Sequence-specific 1H NMR assignments and the collection of conformational constraints were achieved with two-dimensional 1H NMR, and the structure was calculated with the distance geometry program DIANA. The root mean square deviations for the well-defined polypeptide segment of residues 9-34 calculated for the 20 best energy-minimized DIANA conformers relative to their mean are 0.5 and 1.3 A for the polypeptide backbone atoms N, C alpha, and C', and for all heavy atoms, respectively. The 35-residue polypeptide chain of SP-C forms an alpha-helix between positions 9 and 34, which includes two segments of seven and four consecutive valyls that are separated by a single leucyl residue. The N-terminal hexapeptide segment, which includes two palmitoylcysteinyls, is flexibly disordered. The length of the alpha-helix is about 37 A, and the helical segment of residues 13-28, which contains exclusively aliphatic residues with branched side chains, is 23-A long and about 10 A in diameter. The alpha-helix is outstandingly regular, with virtually identical chi 1 angles for all valyl residues. The observation of a helical structure of SP-C was rather unexpected, considering that Val is generally underrepresented in alpha-helices, and it provides intriguing novel insights into the structural basis of SP-C functions as well as into general structural aspects of protein-lipid interactions in biological membranes.
- Institut für Molekularbiologie und Biophysik, Eidenössiche Technische Hochshule-Hönggerberg.
Organizational Affiliation: