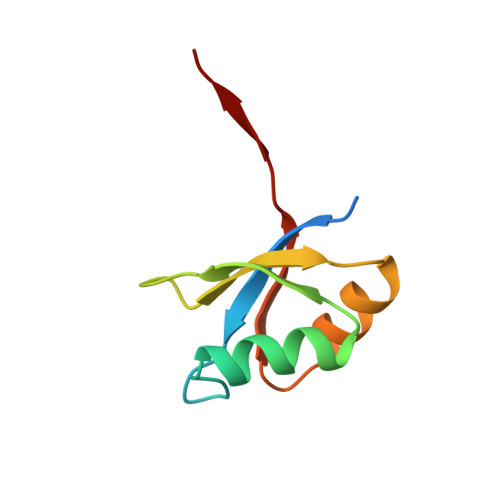
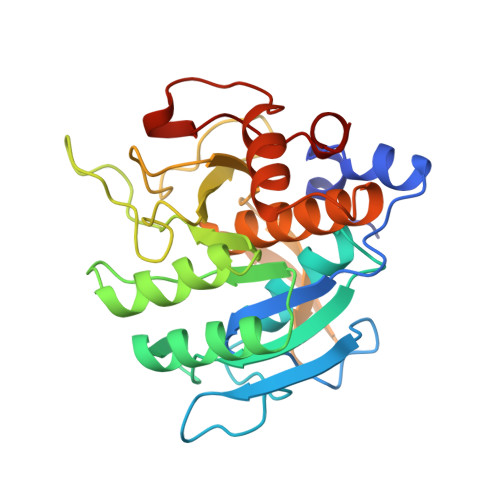
The prosegment-subtilisin BPN' complex: crystal structure of a specific 'foldase'.
Gallagher, T., Gilliland, G., Wang, L., Bryan, P.(1995) Structure 3: 907-914
- PubMed: 8535784
- DOI: https://doi.org/10.1016/S0969-2126(01)00225-8
- Primary Citation of Related Structures:
1SPB - PubMed Abstract:
The folding of the bacterial protease subtilisin BPN' (SBT) is dependent on its 77-residue prosegment, which is then autocatalytically removed to give the mature enzyme. Mature subtilisin represents a class of proteins that lacks an efficient folding pathway. Refolding of mature SBT is extremely slow unless catalyzed by the independently expressed prosegment, leading to a bimolecular complex. We report the crystal structure at 2.0 A resolution of the prosegment-SBT complex and consider its implications for prosubtilisin BPN' maturation and folding catalysis. The prosegment forms a compact domain that binds SBT through an extensive interface involving the enzyme's two parallel surface helices (residues 104-116 and 133-144), supplying negatively charged caps to the N termini of these helices. The prosegment C terminus binds in the enzyme active site in a product-like manner, with Tyr77 in the P1 binding pocket. The structure of the complex supports a unimolecular mechanism for prosubtilisin cleavage, involving a 25 A rearrangement of the SBT N terminus in a late folding step. A mechanism of folding catalysis in which the two helices and their connecting beta strand form a prosegment-stabilized folding nucleus is proposed. While this putative nucleus is stabilized by prosegment binding, the N-terminal and C-terminal subdomains of SBT could fold by propagation.
- Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute, Rockville, USA.
Organizational Affiliation: