Product of enzymatic self-cleavage bound in the active site of HIV protease
Vondrackova, E., Hasek, J., Jaskolski, M., Rezacova, P., Dohnalek, J., Skalova, T., Petrokova, H., Duskova, J., Brynda, J., Sedlacek, J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protease | 99 | Human immunodeficiency virus 1 | Mutation(s): 0 Gene Names: POL EC: 3.4.23.16 | 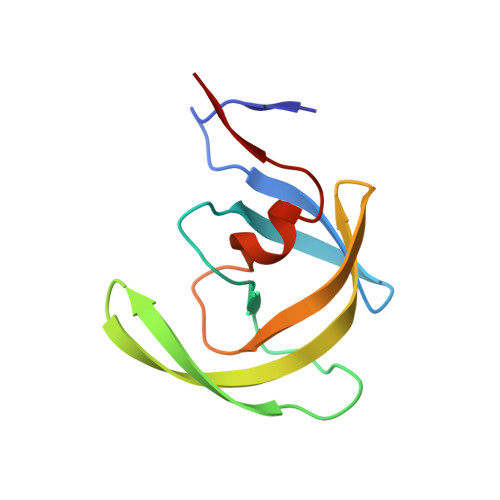 | |
UniProt | |||||
Find proteins for P03367 (Human immunodeficiency virus type 1 group M subtype B (isolate BRU/LAI)) Explore P03367 Go to UniProtKB: P03367 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03367 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5-mer peptide from Protease | C [auth I] | 5 | Human immunodeficiency virus 1 | Mutation(s): 0 Gene Names: POL | 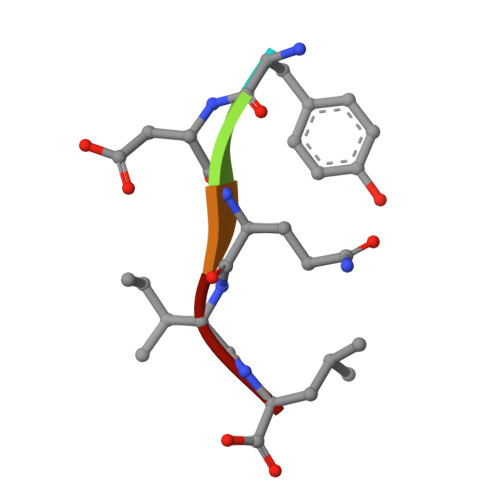 |
UniProt | |||||
Find proteins for P03367 (Human immunodeficiency virus type 1 group M subtype B (isolate BRU/LAI)) Explore P03367 Go to UniProtKB: P03367 | |||||
Entity Groups | |||||
| UniProt Group | P03367 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DMS Query on DMS | H [auth A] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| BME Query on BME | F [auth A], G [auth A], J [auth B] | BETA-MERCAPTOETHANOL C2 H6 O S DGVVWUTYPXICAM-UHFFFAOYSA-N |  | ||
| CL Query on CL | D [auth A], E [auth A], I [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57.765 | α = 90 |
| b = 86.284 | β = 90 |
| c = 45.834 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| CNS | phasing |