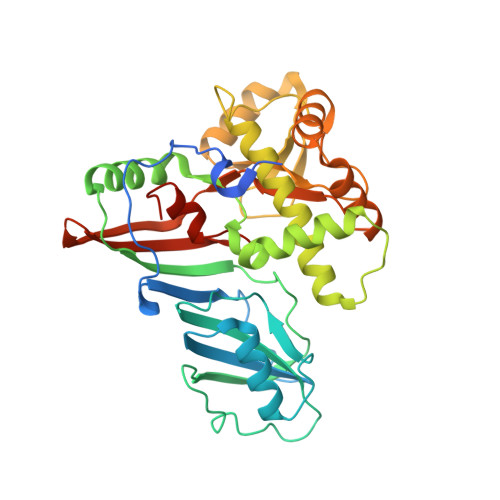
Crystal structure of TruD, a novel pseudouridine synthase with a new protein fold
Kaya, Y., Del Campo, M., Ofengand, J., Malhotra, A.(2004) J Biological Chem 279: 18107-18110
- PubMed: 14999002
- DOI: https://doi.org/10.1074/jbc.C400072200
- Primary Citation of Related Structures:
1SI7 - PubMed Abstract:
TruD, a recently discovered novel pseudouridine synthase in Escherichia coli, is responsible for modifying uridine13 in tRNA(Glu) to pseudouridine. It has little sequence homology with the other 10 pseudouridine synthases in E. coli which themselves have been grouped into four related protein families. Crystal structure determination of TruD revealed a two domain structure consisting of a catalytic domain that differs in sequence but is structurally very similar to the catalytic domain of other pseudouridine synthases and a second large domain (149 amino acids, 43% of total) with a novel alpha/beta fold that up to now has not been found in any other protein.
- Department of Biochemistry and Molecular Biology, University of Miami School of Medicine, Miami, Florida 33101-6129, USA.
Organizational Affiliation: