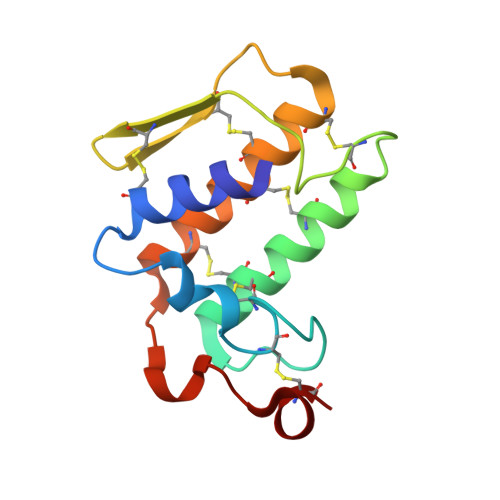
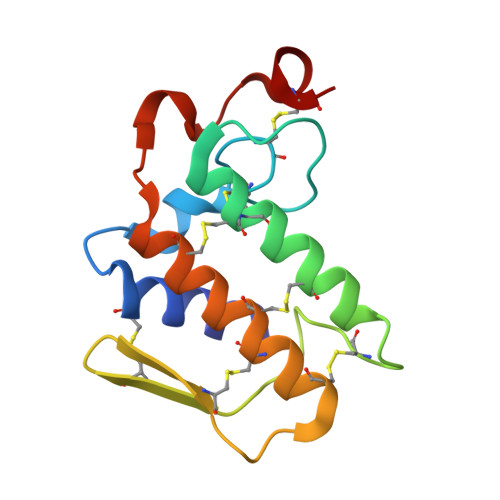
Crystal structure of a calcium-induced dimer of two isoforms of cobra phospholipase A2 at 1.6 A resolution.
Jabeen, T., Sharma, S., Singh, N., Singh, R.K., Kaur, P., Perbandt, M., Betzel, C.h., Srinivasan, A., Singh, T.P.(2005) Proteins 59: 856-863
- PubMed: 15828003
- DOI: https://doi.org/10.1002/prot.20464
- Primary Citation of Related Structures:
1S6B - PubMed Abstract:
The calcium-induced formation of a complex between two isoforms of cobra venom phospholipase A2 reveals a novel interplay between the monomer-dimer and activity-inactivity transitions. The monodispersed isoforms lack activity in the absence of calcium ions while both molecules gain activity in the presence of calcium ions. At concentrations higher than 10 mg/ml, in the presence of calcium ions, they dimerize and lose activity again. The present study reports the crystal structure of a calcium-induced dimer between two isoforms of cobra phospholipase A2. In the complex, one molecule contains a calcium ion in the calcium binding loop while the second molecule does not possess an intramolecular calcium ion. However, there are two calcium ions per dimer in the structure. The second calcium ion is present at an intermolecular site and that is presumably responsible for the dimerization. The calcium binding loops of the two molecules adopt strikingly different conformations. The so-called calcium binding loop in the calcium-containing molecule adopts a normal conformation as generally observed in other calcium containing phospholipase A(2) enzymes while the conformation of the corresponding loop in the calcium free monomer deviates considerably with the formation of a unique intraloop Gly33 (N)-Cys27 (O) = 2.74 A backbone hydrogen bond. The interactions of Arg31 (B) with Asp49 (A) and absence of calcium ion are responsible for the loss of catalytic activity in molecule A while interactions of Arg2 (B) with Tyr52 (B) inactivate molecule B.
- Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.
Organizational Affiliation: