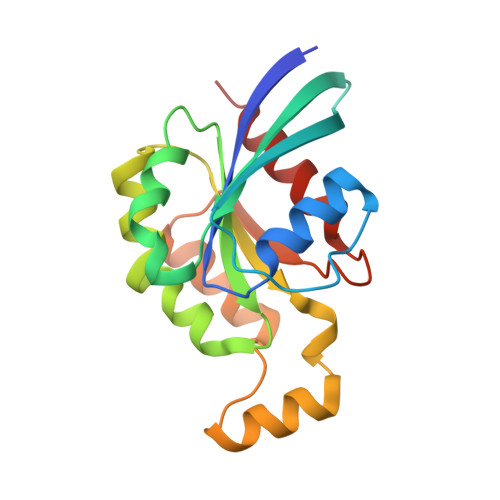
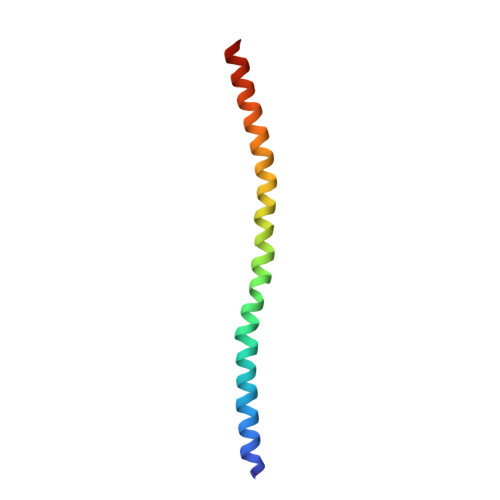Structural Insights into the Interaction of ROCKI with the Switch Regions of RhoA.
Dvorsky, R., Blumenstein, L., Vetter, I.R., Ahmadian, M.R.(2004) J Biological Chem 279: 7098-7104
- PubMed: 14660612
- DOI: https://doi.org/10.1074/jbc.M311911200
- Primary Citation of Related Structures:
1S1C - PubMed Abstract:
The Rho-ROCK pathway modulates the phosphorylation level of a variety of important signaling proteins and is thereby involved in miscellaneous cellular processes including cell migration, neurite outgrowth, and smooth muscle contraction. The observation of the involvement of the Rho-ROCK pathway in tumor invasion and in diseases such as hypertension and bronchial asthma makes it an interesting target for drug development. We herein present the crystal structure of the complex between active RhoA and the Rho-binding domain of ROCKI. The Rho-binding domain structure forms a parallel alpha-helical coiled-coil dimer and, in contrast to the published Rho-protein kinase N structure, binds exclusively to the switch I and II regions of the guanosine 5'-(beta,gamma-imido)triphosphate-bound RhoA. The switch regions of two different RhoA molecules form a predominantly hydrophobic patch, which is complementarily bound by two identical short helices of 13 residues (amino acids 998-1010). The identified ROCK-binding site of RhoA strikingly supports the assumption of a common consensus-binding site for effector recognition.
- Max-Planck-Institute fuer Molekulare Physiologie, Abteilung Strukturelle Biologie, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.
Organizational Affiliation: