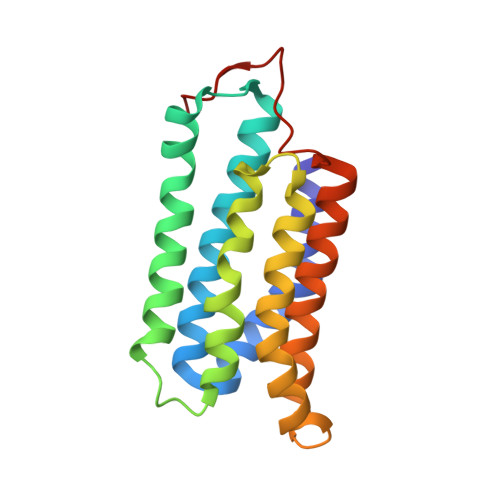
STRUCTURE OF THE N-TERMINAL DOMAIN OF THE ADENYLYL CYCLASE-ASSOCIATED PROTEIN (CAP) FROM DICTYOSTELIUM DISCOIDEUM
Ksiazek, D., Brandstetter, H., Israel, L., Bourenkov, G.P., Katchalova, G., Janssen, K.P., Bartunik, H.D., Noegel, A.A., Schleicher, M., Holak, T.A.(2003) Structure 11: 1171-1178
- PubMed: 12962635
- DOI: https://doi.org/10.1016/s0969-2126(03)00180-1
- Primary Citation of Related Structures:
1S0P - PubMed Abstract:
Cyclase-associated proteins (CAPs) are widely distributed and highly conserved proteins that regulate actin remodeling in response to cellular signals. The N termini of CAPs play a role in Ras signaling and bind adenylyl cyclase; the C termini bind to G-actin and thereby alter the dynamic rearrangements of the microfilament system. We report here the X-ray structure of the core of the N-terminal domain of the CAP from Dictyostelium discoideum, which comprises residues 51-226, determined by a combination of single isomorphous replacement with anomalous scattering (SIRAS). The overall structure of this fragment is an alpha helix bundle composed of six antiparallel helices. Results from gel filtration and crosslinking experiments for CAP(1-226), CAP(255-464), and the full-length protein, together with the CAP N-terminal domain structure and the recently determined CAP C-terminal domain structure, provide evidence that the functional structure of CAP is multimeric.
- Max-Planck Institut für Biochemie, 82152 Martinsried, Germany.
Organizational Affiliation: