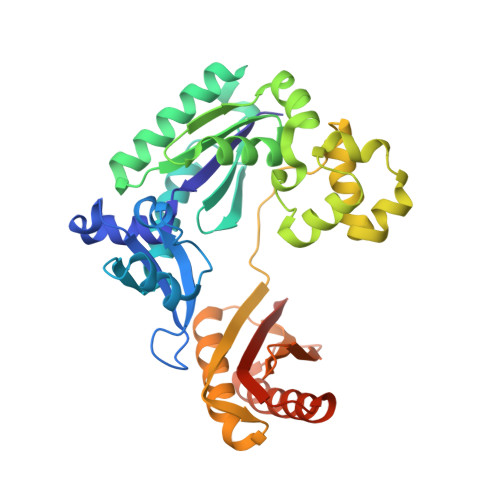
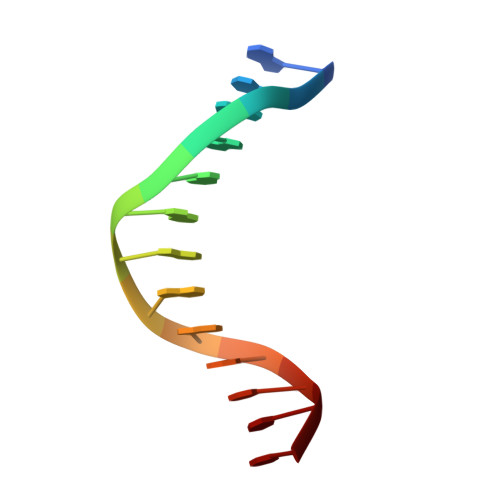
Replication of a Cis-Syn Thymine Dimer at Atomic Resolution
Ling, H., Boudsocq, F., Plosky, B., Woodgate, R., Yang, W.(2003) Nature 424: 1083-1087
- PubMed: 12904819
- DOI: https://doi.org/10.1038/nature01919
- Primary Citation of Related Structures:
1RYR, 1RYS - PubMed Abstract:
Ultraviolet light damages DNA by catalysing covalent bond formation between adjacent pyrimidines, generating cis-syn cyclobutane pyrimidine dimers (CPDs) as the most common lesion. CPDs block DNA replication by high-fidelity DNA polymerases, but they can be efficiently bypassed by the Y-family DNA polymerase pol eta. Mutations in POLH encoding pol eta are implicated in nearly 20% of xeroderma pigmentosum, a human disease characterized by extreme sensitivity to sunlight and predisposition to skin cancer. Here we have determined two crystal structures of Dpo4, an archaeal pol eta homologue, complexed with CPD-containing DNA, where the 3' and 5' thymine of the CPD separately serves as a templating base. The 3' thymine of the CPD forms a Watson-Crick base pair with the incoming dideoxyATP, but the 5' thymine forms a Hoogsteen base pair with the dideoxyATP in syn conformation. Dpo4 retains a similar tertiary structure, but each unusual DNA structure is individually fitted into the active site for catalysis. A model of the pol eta-CPD complex built from the crystal structures of Saccharomyces cerevisiae apo-pol eta and the Dpo4-CPD complex suggests unique features that allow pol eta to efficiently bypass CPDs.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: