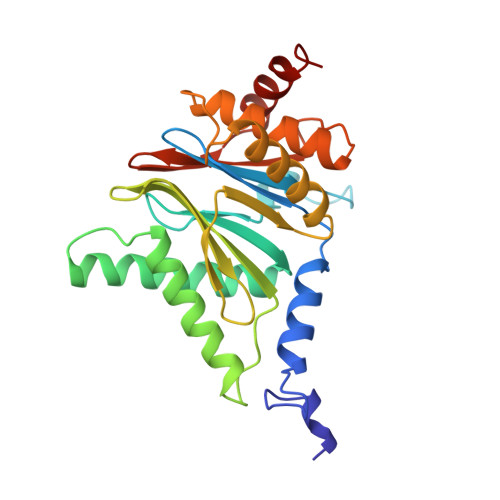
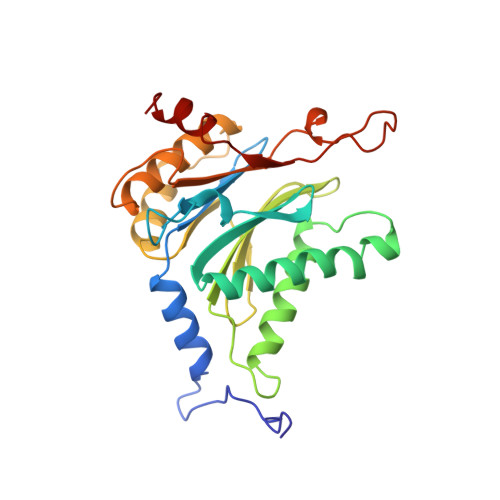
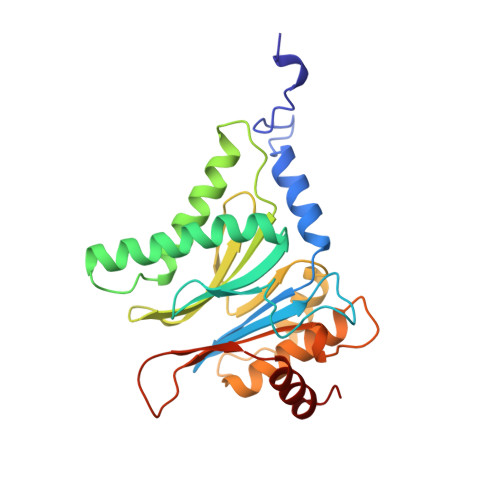
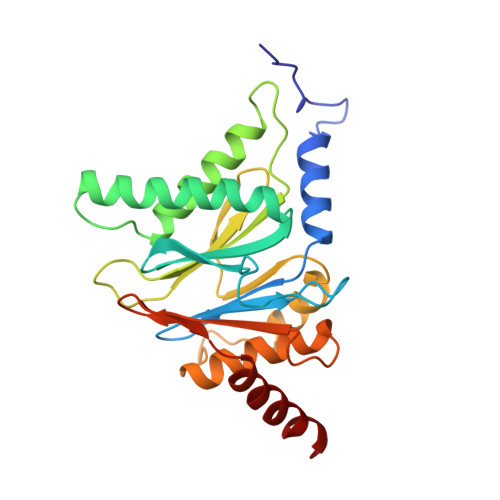
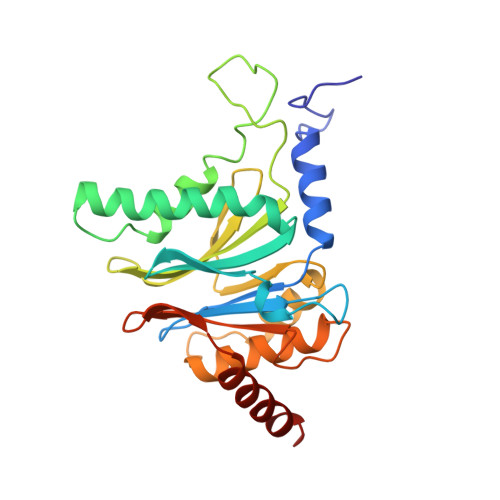
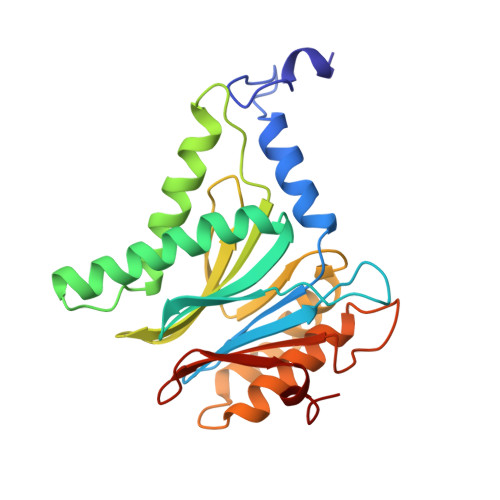
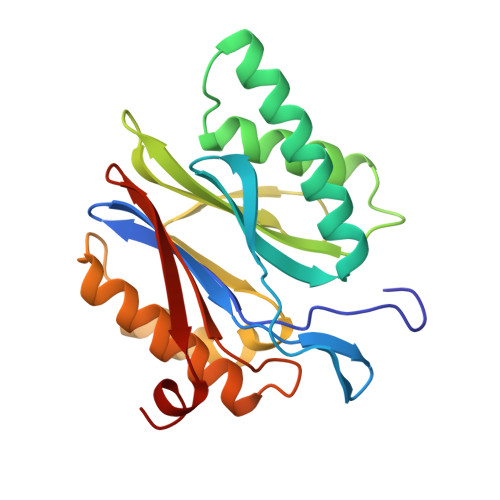
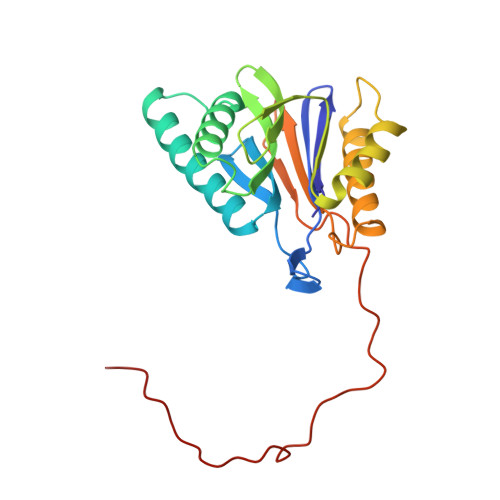
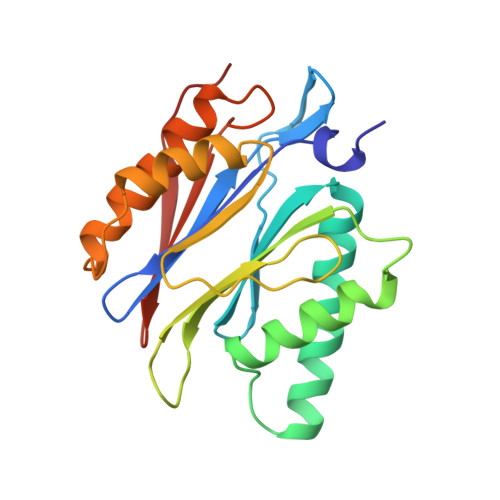
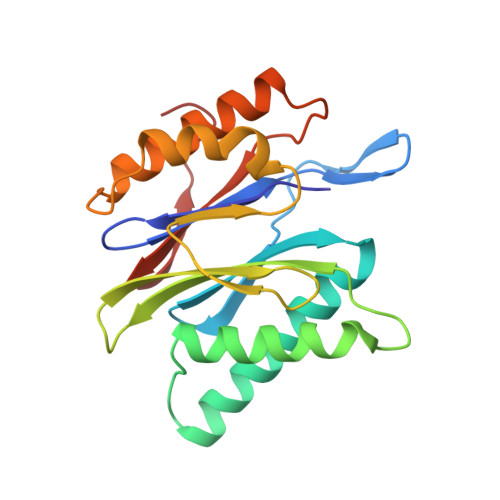
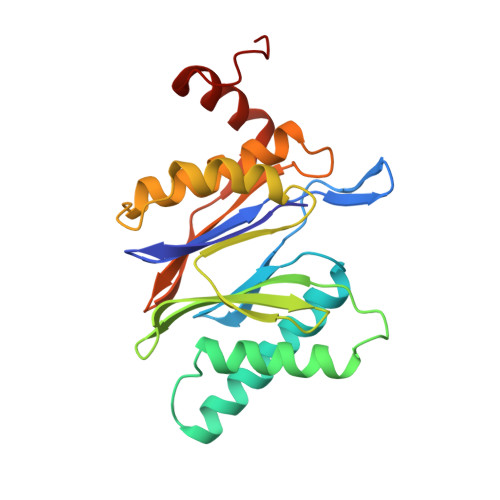
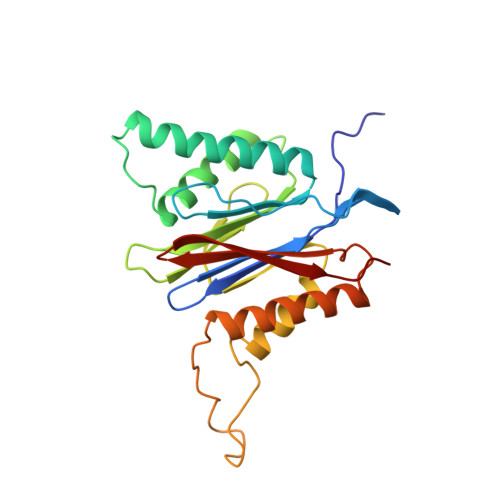
Structure of 20S proteasome from yeast at 2.4 A resolution.
Groll, M., Ditzel, L., Lowe, J., Stock, D., Bochtler, M., Bartunik, H.D., Huber, R.(1997) Nature 386: 463-471
- PubMed: 9087403
- DOI: https://doi.org/10.1038/386463a0
- Primary Citation of Related Structures:
1RYP - PubMed Abstract:
The crystal structure of the 20S proteasome from the yeast Saccharomyces cerevisiae shows that its 28 protein subunits are arranged as an (alpha1...alpha7, beta1...beta7)2 complex in four stacked rings and occupy unique locations. The interior of the particle, which harbours the active sites, is only accessible by some very narrow side entrances. The beta-type subunits are synthesized as proproteins before being proteolytically processed for assembly into the particle. The proforms of three of the seven different beta-type subunits, beta1/PRE3, beta2/PUP1 and beta5/PRE2, are cleaved between the threonine at position 1 and the last glycine of the pro-sequence, with release of the active-site residue Thr 1. These three beta-type subunits have inhibitor-binding sites, indicating that PRE2 has a chymotrypsin-like and a trypsin-like activity and that PRE3 has peptidylglutamyl peptide hydrolytic specificity. Other beta-type subunits are processed to an intermediate form, indicating that an additional nonspecific endopeptidase activity may exist which is important for peptide hydrolysis and for the generation of ligands for class I molecules of the major histocompatibility complex.
- Max-Planck-Institut für Biochemie, Martinsreid, Germany.
Organizational Affiliation: