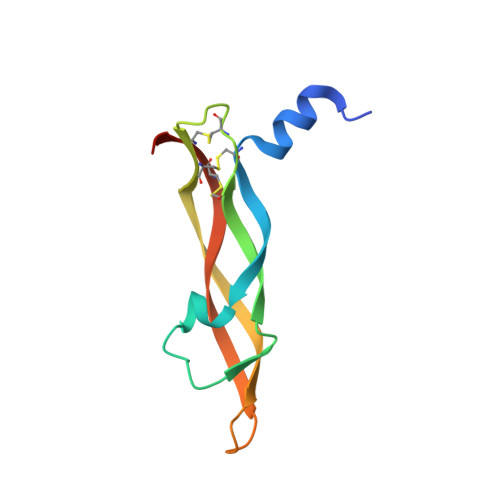
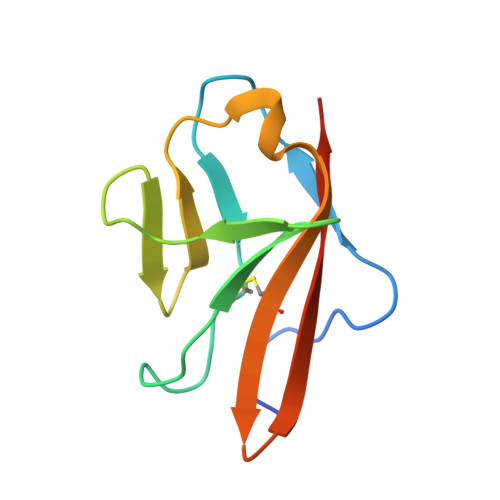
The crystal structure of placental growth factor in complex with domain 2 of vascular endothelial growth factor receptor-1.
Christinger, H.W., Fuh, G., de Vos, A.M., Wiesmann, C.(2004) J Biological Chem 279: 10382-10388
- PubMed: 14684734
- DOI: https://doi.org/10.1074/jbc.M313237200
- Primary Citation of Related Structures:
1RV6 - PubMed Abstract:
Placental growth factor (PlGF) is a member of the vascular endothelial growth factor (VEGF) family and plays an important role in pathological angiogenic events. PlGF exerts its biological activities through binding to VEGFR1, a receptor tyrosine kinase that consists of seven immunoglobulin-like domains in its extracellular portion. Here we report the crystal structure of PlGF bound to the second immunoglobulin-like domain of VEGFR1 at 2.5 A resolution and compare the complex to the closely related structure of VEGF bound to the same receptor domain. The two growth factors, PlGF and VEGF, share a sequence identity of approximately 50%. Despite this moderate sequence conservation, they bind to the same binding interface of VEGFR1 in a very similar fashion, suggesting that both growth factors could induce very similar if not identical signaling events.
- Department of Protein Engineering Genentech, Inc., South San Francisco, California 94080, USA.
Organizational Affiliation: