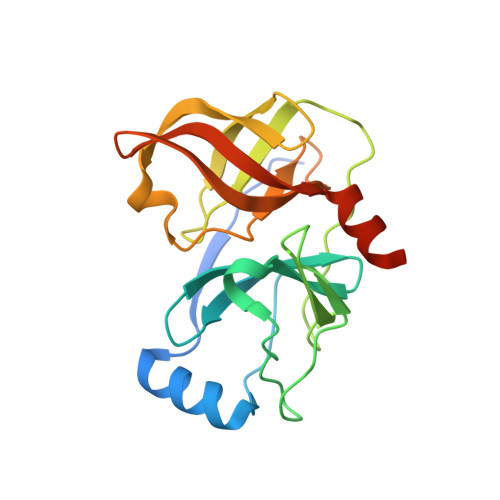
Pyrrolidine-5,5-trans-lactams. 4. Incorporation of a P3/P4 urea leads to potent intracellular inhibitors of hepatitis C virus NS3/4A protease
Slater, M.J., Amphlett, E.M., Andrews, D.M., Bamborough, P., Carey, S.J., Johnson, M.R., Jones, P.S., Mills, G., Parry, N.R., Somers, D.O., Stewart, A.J., Skarzynski, T.(2003) Org Lett 5: 4627-4630
- PubMed: 14627400
- DOI: https://doi.org/10.1021/ol035826v
- Primary Citation of Related Structures:
1RTL - PubMed Abstract:
[reaction: see text] In this, the first of two Letters, we describe how a P3/P4 urea linking unit was used to greatly enhance the biochemical and replicon potency of inhibitors based upon the pyrrolidine-5,5-trans-lactam template. Compound 7b demonstrated a 100 nM IC(50) in the replicon cell-based surrogate HCV assay.
- GlaxoSmithKline Medicines Research Center, Gunnels Wood Road, Stevenage, SG1 2NY, UK. martin.j.slater@gsk.com
Organizational Affiliation: