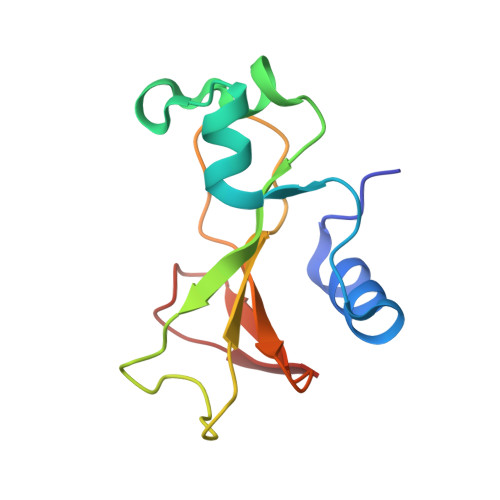
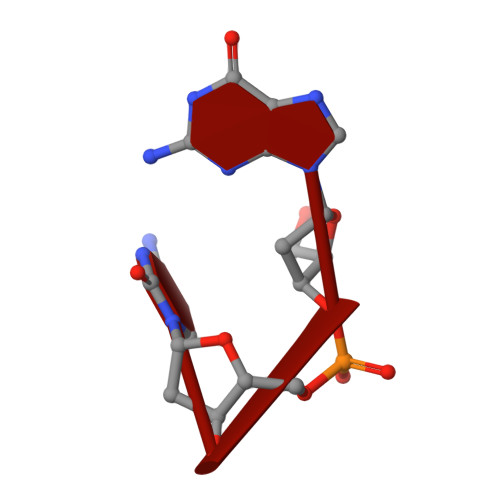
Crystal structure of a barnase-d(GpC) complex at 1.9 A resolution.
Baudet, S., Janin, J.(1991) J Mol Biology 219: 123-132
- PubMed: 2023257
- DOI: https://doi.org/10.1016/0022-2836(91)90862-z
- Primary Citation of Related Structures:
1RNB - PubMed Abstract:
The ribonuclease excreted by Bacillus amyloliquefaciens, Barnase, was co-crystallized with the deoxy-dinucleotide d(GpC). The crystal structure was determined by molecular replacement from a model of free Barnase previously derived by Mauguen et al. Refinement was carried out using data to 1.9 A resolution. The final model, which has a crystallographic R factor of 22%, includes 869 protein atoms, 38 atoms from d(GpC), a sulfate ion and 73 water molecules. Only minor differences from free Barnase are seen in the protein moiety, the root-mean-square C alpha movement being 0.45 A. The dinucleotide has a folded conformation. It is located near the active site of the enzyme, but outside the protein molecule and making crystal packing contacts with neighboring molecules. The guanine base is stacked on the imidazole ring of active site His102, rather than binding to the so-called recognition loop as it does in other complexes of guanine nucleotides with microbial nucleases. The deoxyguanosine is syn, with the sugar ring in C-2'-endo conformation; the deoxycytidine is anti and C-4'-exo. In addition to the stacking interaction, His102 hydrogen bonds to the free 5' hydroxyl, which is located near the position where the 3' phosphate group is found in other inhibitors of microbial ribonucleases. While the mode of binding observed with d(GpC) and Barnase would be non-productive for a dinucleotide substrate, it may define a site for the nucleotide product on the 3' side of the hydrolyzed bond.
- Laboratoire de Biologie Physicochimique-CNRS U.A. 1131, Université Paris-Sud, Orsay, France.
Organizational Affiliation: