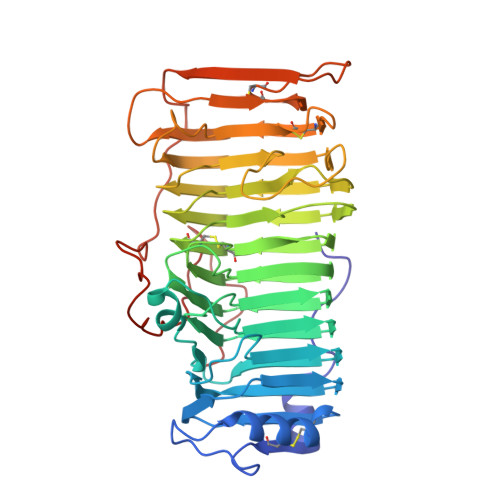
The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel beta helix.
Petersen, T.N., Kauppinen, S., Larsen, S.(1997) Structure 5: 533-544
- PubMed: 9115442
- DOI: https://doi.org/10.1016/s0969-2126(97)00209-8
- Primary Citation of Related Structures:
1RMG - PubMed Abstract:
Pectic substances are the major polysaccharide components of the middle lamella and primary cell wall of dicotyledonous plants. They consist of homogalacturonan 'smooth' regions and highly rhamnified 'hairy' regions of rhamnogalacturonan. The backbone in rhamnogalacturonan-l (RG-l), which is composed of alternating galacturonic acid and rhamnose residues, is the substrate for a new class of enzymes known as rhamnogalacturnoases (RGases). RGase A is a novel enzyme implicated in the enzymatic degradation of RG-l. The structure of RGase A from Aspergillus aculeatus has been solved by the single isomorphous replacement method including anomalous scattering (SIRAS method) to 2.0 A resolution. The enzyme folds into a large right-handed parallel beta helix, with a core composed of 13 turns of beta strands. Four parallel beta sheets (PB1, PB1a, PB2 and PB3), formed by the consecutive turns, are typically separated by a residue in the conformation of a left-handed alpha helix. As a consequence of the consecutive turns, 32% of all residues have their sidechains aligned at the surface or in the interior of the parallel beta helix. The aligned residues at the surface are dominated by threonine, aspartic acid and asparagine, whereas valine, leucine and isoleucine are most frequently found in the interior. A very large hydrophobic cavity is found in the interior of the parallel beta helix. The potential active site is a groove, oriented almost perpendicular to the helical axis, containing a cluster of three aspartic acid residues and one glutamic acid residue. The enzyme is highly glycosylated; two N-linked and eighteen O-linked glycosylation sites have been found in the structure. Rhamnogalacturonase A from A. aculeatus is the first three-dimensional structure of an enzyme hydrolyzing glycoside bonds within the backbone of RG-l. The large groove, which is the potential active site of RGase A, is also seen in the structures of pectate lyases. Two catalytic aspartic acid residues, which have been proposed to have a catalytic role, reside in this area of RGase A. The distance between the aspartic acid residues is consistent with the inverting mechanism of catalysis. The glycan groups bound to RGase A are important to the stability of the crystal, as the carbohydrate moiety is involved in most of the intermolecular hydrogen bonds.
- Centre for Crystallographic Studies, Department of Chemistry, University of Copenhagen, Universitetsparken 5, 2100, Copenhagen, Denmark.
Organizational Affiliation: