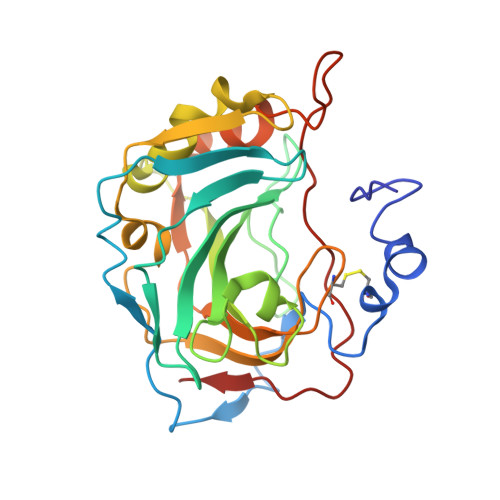
Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes.
Whittington, D.A., Grubb, J.H., Waheed, A., Shah, G.N., Sly, W.S., Christianson, D.W.(2004) J Biological Chem 279: 7223-7228
- PubMed: 14660577
- DOI: https://doi.org/10.1074/jbc.M310809200
- Primary Citation of Related Structures:
1RJ5, 1RJ6 - PubMed Abstract:
Carbonic anhydrase (CA) XIV is the most recently identified mammalian carbonic anhydrase isozyme, and its presence has been demonstrated in a number of tissues. Full-length CA XIV is a transmembrane protein composed of an extracellular catalytic domain, a single transmembrane helix, and a short intracellular polypeptide segment. The amino acid sequence identity of human CA XIV relative to the other membrane-associated isozymes (CA IV, CA IX, and CA XII) is 34-46%. We report here the expression and purification of both the full-length enzyme and a truncated, secretory form of murine CA XIV. Both forms of this isozyme are highly active, and both show an abrogation of activity in the presence of 0.2% SDS, in contrast to the behavior of murine CA IV. We also report the crystal structure of the extracellular domain of murine CA XIV at 2.8 A resolution and of an enzyme-acetazolamide complex at 2.9 A resolution. The structure shows a monomeric glycoprotein with a topology similar to that of other mammalian CA isozymes. Based on the x-ray crystallographic results, we compare and contrast known structures of membrane-associated CA isozymes to rationalize the structural elements responsible for the SDS resistance of CA IV and to discuss prospects for the design of selective inhibitors of membrane-associated CA isozymes.
- Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA.
Organizational Affiliation: