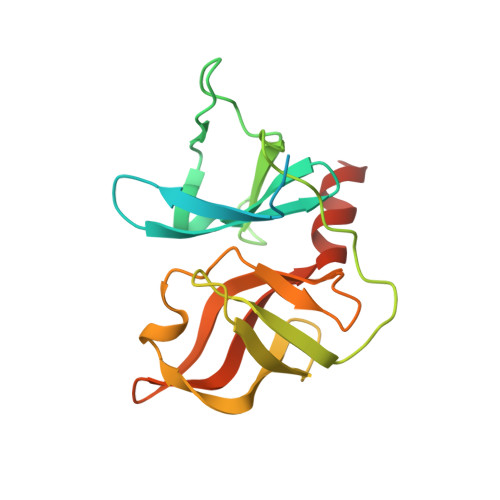
Hepatitis C NS3 protease inhibition by peptidyl-alpha-ketoamide inhibitors: kinetic mechanism and structure.
Liu, Y., Stoll, V.S., Richardson, P.L., Saldivar, A., Klaus, J.L., Molla, A., Kohlbrenner, W., Kati, W.M.(2004) Arch Biochem Biophys 421: 207-216
- PubMed: 14984200
- DOI: https://doi.org/10.1016/j.abb.2003.11.013
- Primary Citation of Related Structures:
1RGQ - PubMed Abstract:
A series of novel peptidyl-alpha-ketoamide compounds were evaluated as inhibitors of the deltaNS3-NS4A serine protease from the hepatitis C virus. These peptidyl-alpha-ketoamide inhibitors with Ki values ranging from 0.17 nM to 5.6 microM exhibited slow-binding inhibition. Kinetic studies established one-step kinetic mechanisms and dissociation rate constants in the 3-7 x 10(-5) s(-1) range for these compounds. The association rate constants, which ranged from 10 to 263,000 M(-1) s(-1), were responsible for the greater than four order of magnitude overall binding affinity range exhibited by this series. An X-ray crystal structure of a protease-inhibitor complex revealed an unusual interaction between the oxyanion of the adduct and the protein as well as a significant movement in the S1' region of the protein loop comprising residues 35-42. These results are quite different from peptidyl-alpha-ketoacid inhibition of HCV protease, which reportedly undergoes no notable conformational changes and proceeds with a two-step slow-binding kinetic mechanism.
- Antiviral Research, Infectious Disease Research and Advanced Technology, Global Pharmaceutical Research and Development, Abbott Laboratories, Abbott Park, IL 60064-6217, USA. yaya.liu@abbott.com
Organizational Affiliation: