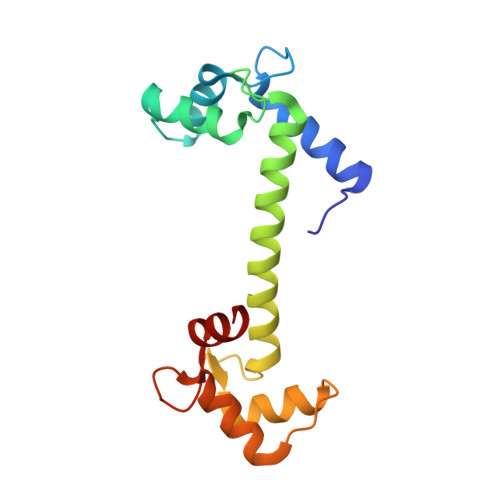
Structure of potato calmodulin PCM6: the first report of the three-dimensional structure of a plant calmodulin.
Yun, C.H., Bai, J., Sun, D.Y., Cui, D.F., Chang, W.R., Liang, D.C.(2004) Acta Crystallogr D Biol Crystallogr 60: 1214-1219
- PubMed: 15213382
- DOI: https://doi.org/10.1107/S0907444904009771
- Primary Citation of Related Structures:
1RFJ - PubMed Abstract:
The crystal structure of a potato calmodulin (PCM6) was solved by molecular replacement and refined to a crystallographic R factor of 22.8% (R(free) = 25.0%) using X-ray diffraction data in the resolution range 8.0-2.0 A. This is the first report of the three-dimensional structure of a plant Ca(2+)-calmodulin. PCM6 crystallizes in a crystal form that belongs to space group P2(1)2(1)2(1), which is different to that of most other calmodulin crystals. The main structural difference between PCM6 and the other calmodulins is in the central helix region and appears to be caused by crystal packing. The surface properties of PCM6 molecules were compared with those of animal calmodulins, which provided an explanation for the unique crystal-packing state of PCM6.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.
Organizational Affiliation: