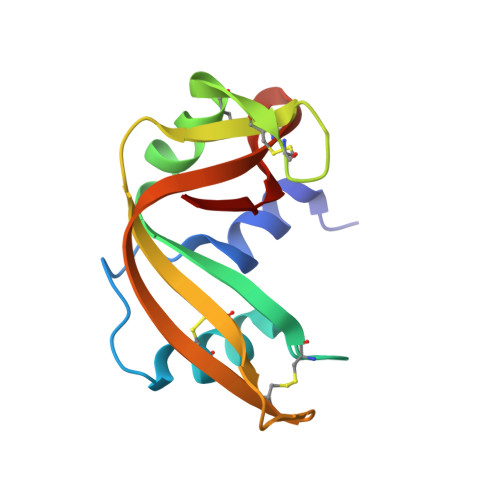
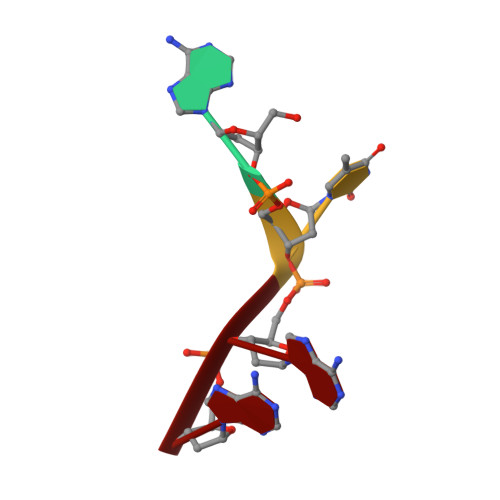
Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition.
Fontecilla-Camps, J.C., de Llorens, R., le Du, M.H., Cuchillo, C.M.(1994) J Biological Chem 269: 21526-21531
- PubMed: 8063789
- DOI: https://doi.org/10.2210/pdb1rcn/pdb
- Primary Citation of Related Structures:
1RCN - PubMed Abstract:
The crystal structure of the complex between ribonuclease A and d(ApTpApApG) has been solved by x-ray crystallography using the molecular replacement method. The model includes, besides the enzyme, the d(ApTpApA) 5'-segment (A1T2A3A4) and 68 solvent molecules. The R-factor for the strongest 87% of the measured data that partially extends to 2.3-A resolution is 0.207. The A1 position is well defined; the 5'-O of the deoxyribose establishes a hydrogen bond with a solvent molecule that is, in turn, bonded to the epsilon-amino group of Lys66. The base (B0 site) is well ordered; it interacts with a symmetry-related enzyme molecule. In the crystal, the phosphate group at the p0 site has no direct charge compensation. However, Lys66 is not too far, and, in solution, it could bind to it. The T2 (R1B1p1) site is occupied as in other complex structures, and it is defined by very good electron density. The A3 site shows that the adenine moiety interacts with Asn71 and Gln69 and that the phosphate at p2 forms a salt bridge with Lys7. The most consistent model for the base of A4 (B3), both in terms of electron density and stereochemistry, shows that it forms a hydrogen bond with Gln69 and a g-g- array with the base at B2. The stacking of B2 and B3 may be a general feature of the binding of polyribonucleotides to ribonuclease A. The side chains of Gln69, Asn71, and Glu111 may thus constitute a malleable binding site capable of establishing various hydrogen bonds depending on the nature of the stacked bases. There is no evidence for the 3' G5 site in the electron density map.
- Laboratoire de Cristallographie et de Cristallogénèse des Protéines, Institut de Biologie Structurale, Grenoble, France.
Organizational Affiliation: