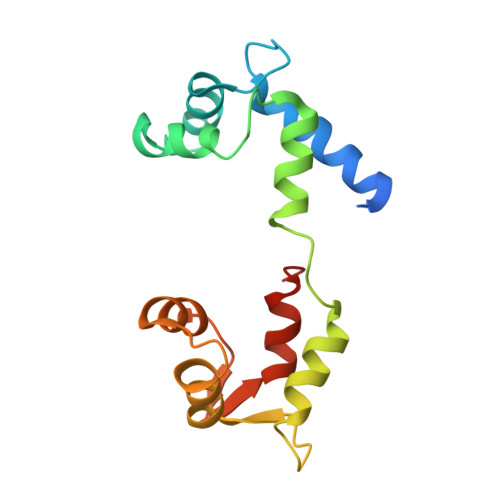
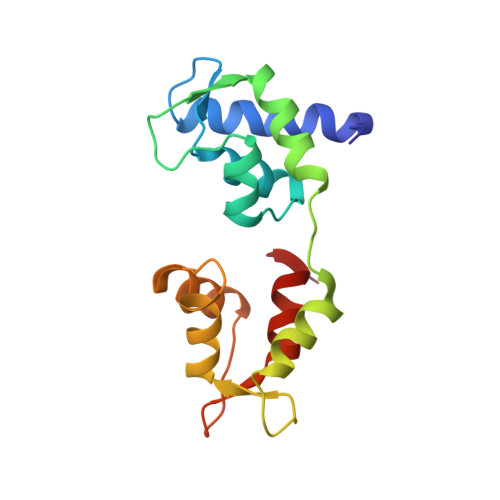
Crystal structure of scallop Myosin s1 in the pre-power stroke state to 2.6 a resolution: flexibility and function in the head.
Gourinath, S., Himmel, D.M., Brown, J.H., Reshetnikova, L., Szent-Gyorgyi, A.G., Cohen, C.(2003) Structure 11: 1621-1627
- PubMed: 14656445
- DOI: https://doi.org/10.1016/j.str.2003.10.013
- Primary Citation of Related Structures:
1QVI - PubMed Abstract:
We have extended the X-ray structure determination of the complete scallop myosin head in the pre-power stroke state to 2.6 A resolution, allowing an atomic comparison of the three major (weak actin binding) states of various myosins. We can now account for conformational differences observed in crystal structures in the so-called "pliant region" at the motor domain-lever arm junction between scallop and vertebrate smooth muscle myosins. A hinge, which may contribute to the compliance of the myosin crossbridge, has also been identified for the first time within the regulatory light-chain domain of the lever arm. Analysis of temperature factors of key joints of the motor domain, especially the SH1 helix, provides crystallographic evidence for the existence of the "internally uncoupled" state in diverse isoforms. The agreement between structural and solution studies reinforces the view that the unwinding of the SH1 helix is a part of the cross-bridge cycle in many myosins.
- Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, MA 02454, USA.
Organizational Affiliation: