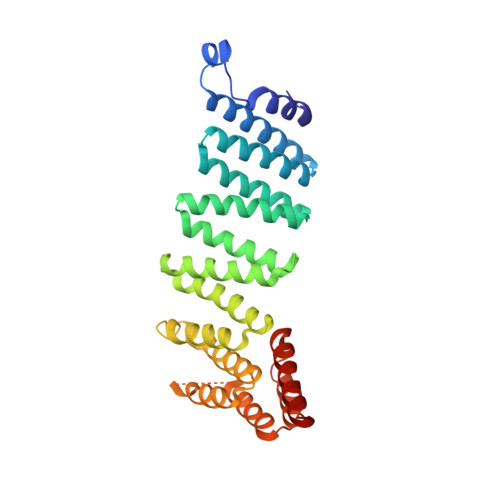
Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly.
Rice, L.M., Brunger, A.T.(1999) Mol Cell 4: 85-95
- PubMed: 10445030
- DOI: https://doi.org/10.1016/s1097-2765(00)80190-2
- Primary Citation of Related Structures:
1QQE - PubMed Abstract:
SNAP proteins play an essential role in membrane trafficking in eukaryotic cells. They activate and recycle SNARE proteins by serving as adaptors between SNAREs and the cytosolic chaperone NSF. We have determined the crystal structure of Sec17, the yeast homolog of alpha-SNAP, to 2.9 A resolution. Sec17 is composed of an N-terminal twisted sheet of alpha-helical hairpins and a C-terminal alpha-helical bundle. The N-terminal sheet has local similarity to the tetratricopeptide repeats from protein phosphatase 5 but has a different overall twist. Sec17 also shares structural features with HEAT and clathrin heavy chain repeats. Possible models of SNAP:SNARE binding suggest that SNAPs may function as lever arms, transmitting forces generated by conformational changes in NSF/Sec18 to drive disassembly of SNARE complexes.
- Howard Hughes Medical Institute, Yale University, New Haven, Connecticut 06520, USA.
Organizational Affiliation: