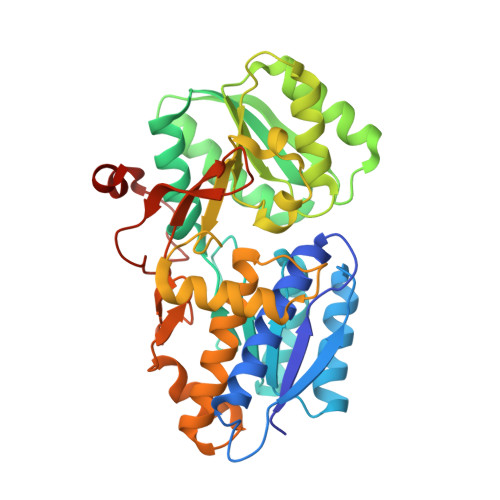
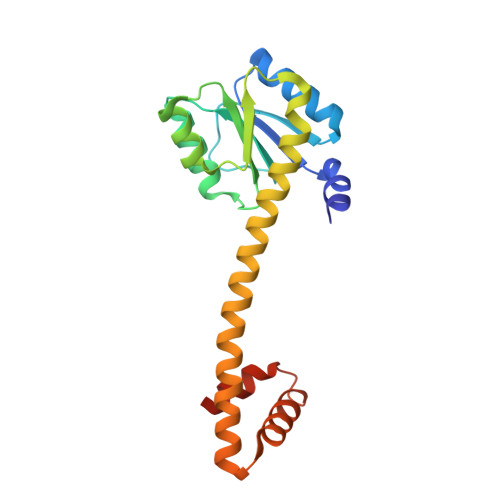
Crystal Structure and Induction Mechanism of Amic-Amir: A Ligand-Regulated Transcription Antitermination Complex
O'Hara, B.P., Norman, R.A., Wan, P.T.C., Roe, S.M., Barrett, T.E., Drew, R.E., Pearl, L.H.(1999) EMBO J 18: 5175
- PubMed: 10508151
- DOI: https://doi.org/10.1093/emboj/18.19.5175
- Primary Citation of Related Structures:
1QO0 - PubMed Abstract:
Inducible expression of the aliphatic amidase operon in Pseudomonas aeruginosa is controlled by an antitermination mechanism which allows production of the full-length transcript only in the presence of small-molecule inducers, such as acetamide. Ligand-regulated antitermination is provided by AmiC, the ligand-sensitive negative regulator, and AmiR, the RNA-binding positive regulator. Under non-inducing or repressing growth conditions, AmiC and AmiR form a complex in which the activity of AmiR is silenced. The crystal structure of the AmiC-AmiR complex identifies AmiR as a new and highly unusual member of the response-regulator family of bacterial signal transduction proteins, regulated by sequestration rather than phosphorylation. Comparison with the structure of free AmiC reveals the subtle mechanism of ligand-induced release of AmiR.
- Centre for Structural Biology, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, UK.
Organizational Affiliation: