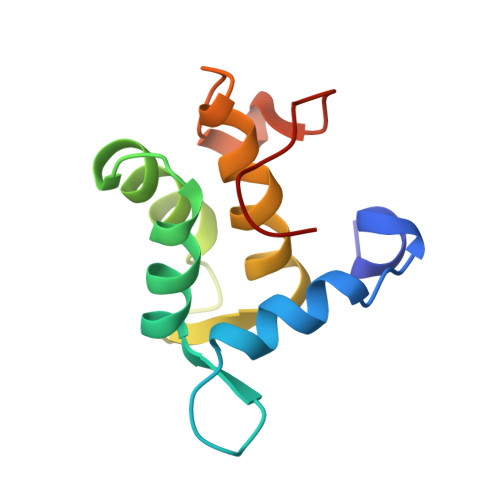
The Eh1 Domain of Eps15 is Structurally Classified as a Member of the S100 Subclass of EF-Hand Containing Proteins
Whitehead, B., Tessari, M., Carotenuto, A., van Bergen en Henegouwen, P.M., Vuister, G.W.(1999) Biochemistry 38: 11271-11277
- PubMed: 10471276
- DOI: https://doi.org/10.1021/bi990922i
- Primary Citation of Related Structures:
1QJT - PubMed Abstract:
The Eps15 homology (EH) domain is a protein-protein interaction module that binds to proteins containing the asparagine-proline-phenylalanine (NPF) or tryptophan/phenylalanine-tryptophan (W/FW) motif. EH domain-containing proteins serve important roles in signaling and processes connected to transport, protein sorting, and organization of subcellular structure. Here, we report the solution structure of the apo form of the EH1 domain of mouse Eps15, as determined by high-resolution multidimensional heteronuclear NMR spectroscopy. The polypeptide folds into six alpha-helices and a short antiparallel beta-sheet. Additionally, it contains a long, structured, topologically unique C-terminal loop. Helices 2-5 form two EF-hand motifs. Structural similarity and Ca(2+) binding properties lead to classification of the EH1 domain as a member of the S100 subclass of EF-hand-containing proteins, albeit with a unique set of interhelical angles. Binding studies using an eight-residue NPF-containing peptide derived from RAB, the cellular cofactor of the HIV Rev protein, show a hydrophobic peptide-binding pocket formed by conserved tryptophan and leucine residues.
- Nijmegen NSR Center for Molecular Structure, Design and Synthesis, Laboratory of Biophysical Chemistry, University of Nijmegen, The Netherlands.
Organizational Affiliation: