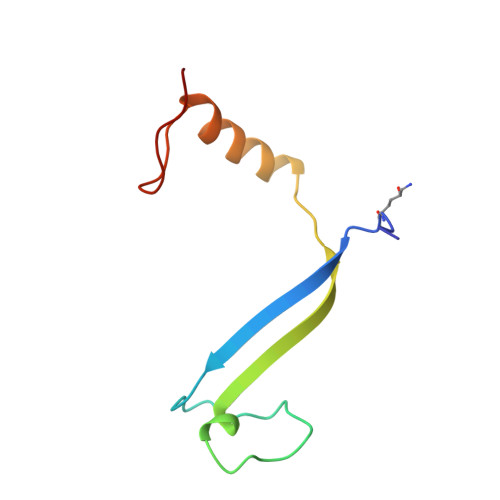
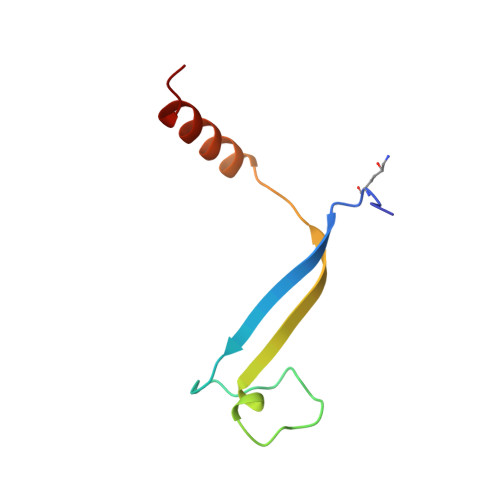
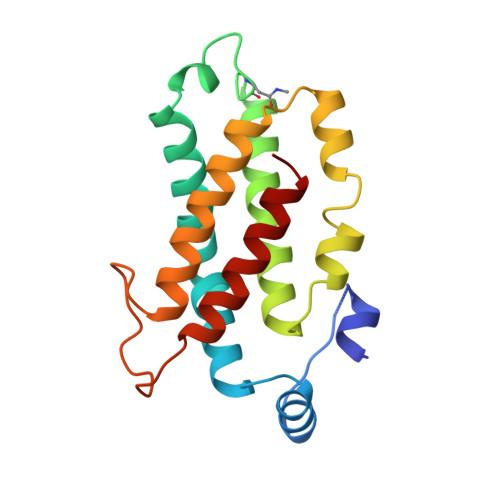
Evolution of a light-harvesting protein by addition of new subunits and rearrangement of conserved elements: crystal structure of a cryptophyte phycoerythrin at 1.63-A resolution.
Wilk, K.E., Harrop, S.J., Jankova, L., Edler, D., Keenan, G., Sharples, F., Hiller, R.G., Curmi, P.M.(1999) Proc Natl Acad Sci U S A 96: 8901-8906
- PubMed: 10430868
- DOI: https://doi.org/10.1073/pnas.96.16.8901
- Primary Citation of Related Structures:
1QGW - PubMed Abstract:
Cryptophytes are unicellular photosynthetic algae that use a lumenally located light-harvesting system, which is distinct from the phycobilisome structure found in cyanobacteria and red algae. One of the key components of this system is water-soluble phycoerythrin (PE) 545 whose expression is enhanced by low light levels. The crystal structure of the heterodimeric alpha(1)alpha(2)betabeta PE 545 from the marine cryptophyte Rhodomonas CS24 has been determined at 1.63-A resolution. Although the beta-chain structure is similar to the alpha and beta chains of other known phycobiliproteins, the overall structure of PE 545 is novel with the alpha chains forming a simple extended fold with an antiparallel beta-ribbon followed by an alpha-helix. The two doubly linked beta50/beta61 chromophores (one on each beta subunit) are in van der Waals contact, suggesting that exciton-coupling mechanisms may alter their spectral properties. Each alpha subunit carries a covalently linked 15,16-dihydrobiliverdin chromophore that is likely to be the final energy acceptor. The architecture of the heterodimer suggests that PE 545 may dock to an acceptor protein via a deep cleft and that energy may be transferred via this intermediary protein to the reaction center.
- Initiative in Biomolecular Structure, School of Physics, University of New South Wales, Sydney, NSW 2052, Australia.
Organizational Affiliation: