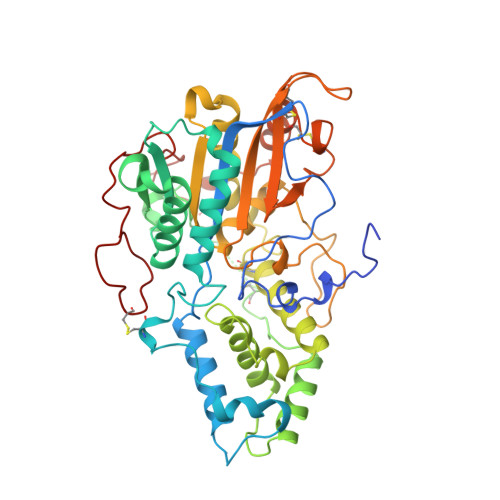
Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2. 4 A resolution.
Kostrewa, D., Wyss, M., D'Arcy, A., van Loon, A.P.(1999) J Mol Biology 288: 965-974
- PubMed: 10329192
- DOI: https://doi.org/10.1006/jmbi.1999.2736
- Primary Citation of Related Structures:
1QFX - PubMed Abstract:
The crystal structure of Aspergillus niger pH 2.5 acid phosphatase (EC 3.1.3.2) has been determined at 2.4 A resolution. In the crystal, two dimers form a tetramer in which the active sites are easily accessible to substrates. The main contacts in the dimer come from the N termini, each lying on the surface of the neighbouring molecule. The monomer consists of two domains, with the active site located at their interface. The active site has a highly conserved catalytic center and a charge distribution, which explains the highly acidic pH optimum and the broad substrate specificity of the enzyme.
- F. Hoffmann-La Roche Ltd, B/65/R312, Basel, 4070, Switzerland. dirk.kostrewa@roche.com
Organizational Affiliation: