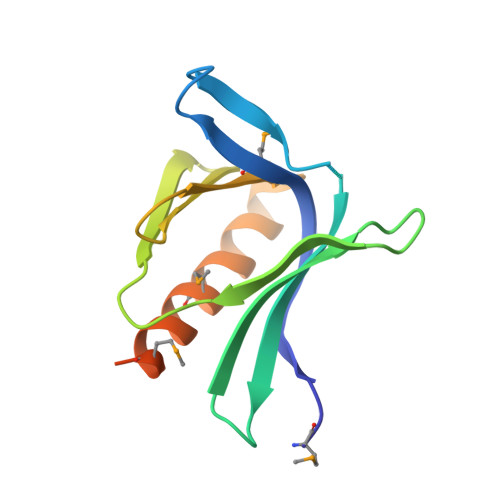
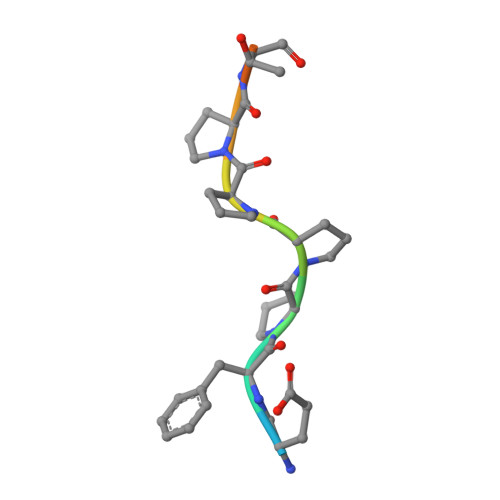
Structure of EVH1, a novel proline-rich ligand-binding module involved in cytoskeletal dynamics and neural function
Fedorov, A.A., Fedorov, E., Gertler, F., Almo, S.C.(1999) Nat Struct Biol 6: 661-665
- PubMed: 10404224
- DOI: https://doi.org/10.1038/10717
- Primary Citation of Related Structures:
1QC6 - PubMed Abstract:
The Ena-VASP homology (EVH1) domain is a protein interaction module found in several proteins that are involved in transducing migratory and morphological signals into cytoskeletal reorganization. EVH1 specifically recognizes proline-rich sequences in its binding partners and directs the localization and formation of multicomponent assemblies involved in actin-based motile processes and neural development. The structure of the complex between an EVH1 domain and the target peptide sequence EFPPPPT identifies the interactions responsible for recognition and distinguishes it from other proline-rich binding modules, including SH3 and WW domains. Surprisingly, the EVH1 domain has structural similarity to pleckstrin homology (PH), phosphotyrosine-binding (PTB) and ran-binding (RanBD) domains.
- Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461, USA.
Organizational Affiliation: