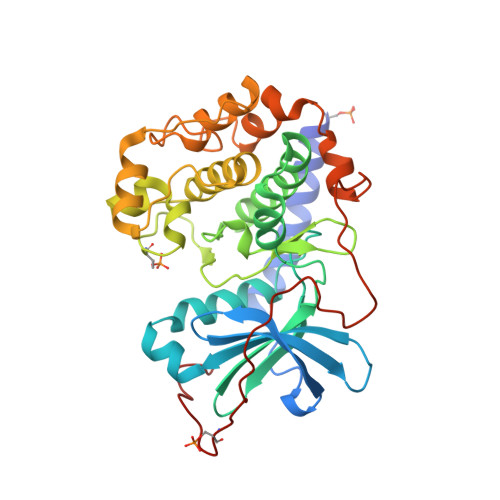
Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity.
Breitenlechner, C., Gassel, M., Hidaka, H., Kinzel, V., Huber, R., Engh, R.A., Bossemeyer, D.(2003) Structure 11: 1595-1607
- PubMed: 14656443
- DOI: https://doi.org/10.1016/j.str.2003.11.002
- Primary Citation of Related Structures:
1Q8T, 1Q8U, 1Q8W - PubMed Abstract:
Protein kinases require strict inactivation to prevent spurious cellular signaling; overactivity can cause cancer or other diseases and necessitates selective inhibition for therapy. Rho-kinase is involved in such processes as tumor invasion, cell adhesion, smooth muscle contraction, and formation of focal adhesion fibers, as revealed using inhibitor Y-27632. Another Rho-kinase inhibitor, HA-1077 or Fasudil, is currently used in the treatment of cerebral vasospasm; the related nanomolar inhibitor H-1152P improves on its selectivity and potency. We have determined the crystal structures of HA-1077, H-1152P, and Y-27632 in complexes with protein kinase A (PKA) as a surrogate kinase to analyze Rho-kinase inhibitor binding properties. Features conserved between PKA and Rho-kinase are involved in the key binding interactions, while a combination of residues at the ATP binding pocket that are unique to Rho-kinase may explain the inhibitors' Rho-kinase selectivity. Further, a second H-1152P binding site potentially points toward PKA regulatory domain interaction modulators.
- Abteilung Strukturforschung, Max-Planck-Institut fuer Biochemie, 82152 Martinsried, Germany.
Organizational Affiliation: