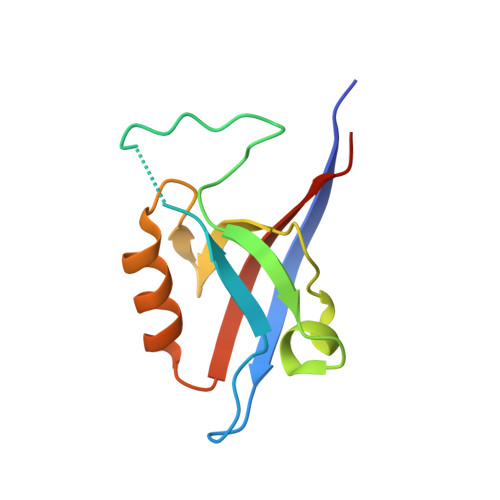
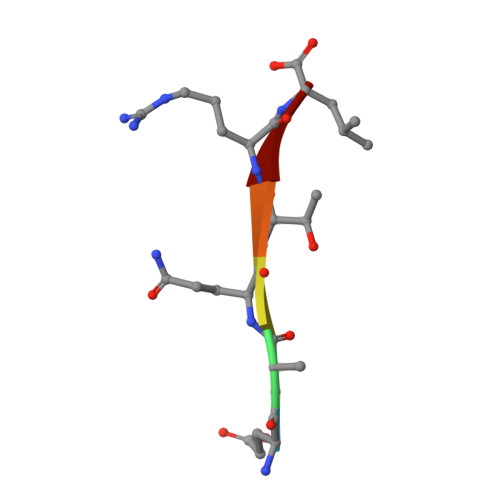
Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
Im, Y.J., Lee, J.H., Park, S.H., Park, S.J., Rho, S.-H., Kang, G.B., Kim, E., Eom, S.H.(2003) J Biological Chem 278: 48099-48104
- PubMed: 12954649
- DOI: https://doi.org/10.1074/jbc.M306919200
- Primary Citation of Related Structures:
1Q3O, 1Q3P - PubMed Abstract:
The Shank/proline-rich synapse-associated protein family of multidomain proteins is known to play an important role in the organization of synaptic multiprotein complexes. For instance, the Shank PDZ domain binds to the C termini of guanylate kinase-associated proteins, which in turn interact with the guanylate kinase domain of postsynaptic density-95 scaffolding proteins. Here we describe the crystal structures of Shank1 PDZ in its peptide free form and in complex with the C-terminal hexapeptide (EAQTRL) of guanylate kinase-associated protein (GKAP1a) determined at 1.8- and 2.25-A resolutions, respectively. The structure shows the typical class I PDZ interaction of PDZ-peptide complex with the consensus sequence -X-(Thr/Ser)-X-Leu. In addition, Asp-634 within the Shank1 PDZ domain recognizes the positively charged Arg at -1 position and hydrogen bonds, and salt bridges between Arg-607 and the side chains of the ligand at -3 and -5 positions contribute further to the recognition of the peptide ligand. Remarkably, whether free or complexed, Shank1 PDZ domains form dimers with a conserved beta B/beta C loop and N-terminal beta A strands, suggesting a novel model of PDZ-PDZ homodimerization. This implies that antiparallel dimerization through the N-terminal beta A strands could be a common configuration among PDZ dimers. Within the dimeric structure, the two-peptide binding sites are arranged so that the N termini of the bound peptide ligands are in close proximity and oriented toward the 2-fold axis of the dimer. This configuration may provide a means of facilitating dimeric organization of PDZ-target assemblies.
- Department of Life Science, Kwangju Institute of Science and Technology, Gwangju 500-712, South Korea.
Organizational Affiliation: