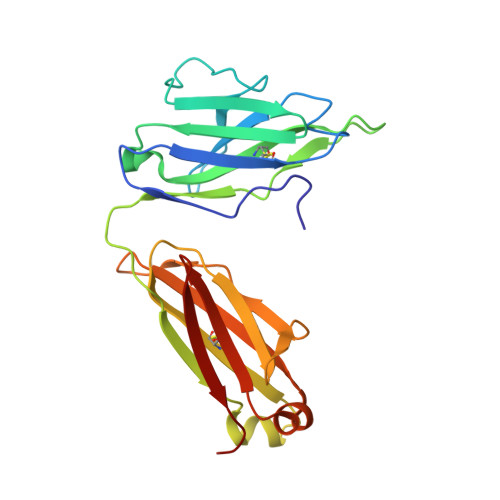
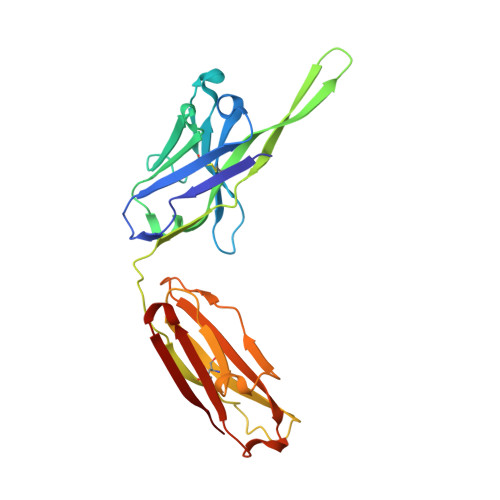
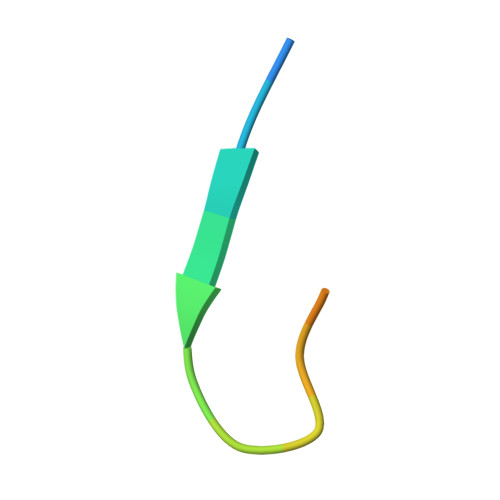
Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D.
Stanfield, R.L., Gorny, M.K., Williams, C., Zolla-Pazner, S., Wilson, I.A.(2004) Structure 12: 193-204
- PubMed: 14962380
- DOI: https://doi.org/10.1016/j.str.2004.01.003
- Primary Citation of Related Structures:
1Q1J - PubMed Abstract:
447-52D is a human monoclonal antibody isolated from a heterohybridoma derived from an HIV-1-infected individual. This antibody recognizes the hypervariable gp120 V3 loop, and neutralizes both X4 and R5 primary isolates, making it one of the most effective anti-V3 antibodies characterized to date. The crystal structure of the 447-52D Fab in complex with a 16-mer V3 peptide at 2.5 A resolution reveals that the peptide beta hairpin forms a three-stranded mixed beta sheet with complementarity determining region (CDR) H3, with most of the V3 side chains exposed to solvent. Sequence specificity is conferred through interaction of the type-II turn (residues GPGR) at the apex of the V3 hairpin with the base of CDR H3. This novel mode of peptide-antibody recognition enables the antibody to bind to many different V3 sequences where only the GPxR core epitope is absolutely required.
- Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: