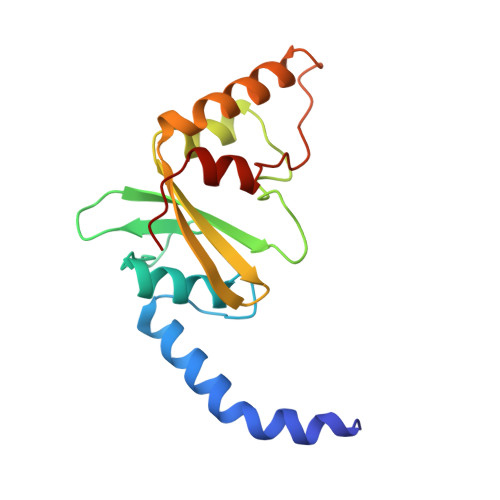
Structure of PvuII endonuclease with cognate DNA.
Cheng, X., Balendiran, K., Schildkraut, I., Anderson, J.E.(1994) EMBO J 13: 3927-3935
- PubMed: 8076590
- DOI: https://doi.org/10.1002/j.1460-2075.1994.tb06708.x
- Primary Citation of Related Structures:
1PVI - PubMed Abstract:
We have determined the structure of PvuII endonuclease complexed with cognate DNA by X-ray crystallography. The DNA substrate is bound with a single homodimeric protein, each subunit of which reveals three structural regions. The catalytic region strongly resembles structures of other restriction endonucleases, even though these regions have dissimilar primary sequences. Comparison of the active site with those of EcoRV and EcoRI endonucleases reveals a conserved triplet sequence close to the reactive phosphodiester group and a conserved acidic pair that may represent the ligands for the catalytic cofactor Mg2+. The DNA duplex is not significantly bent and maintains a B-DNA-like conformation. The subunit interface region of the homodimeric protein consists of a pseudo-three-helix bundle. Direct contacts between the protein and the base pairs of the PvuII recognition site occur exclusively in the major groove through two antiparallel beta strands from the sequence recognition region of the protein. Water-mediated contacts are made in the minor grooves to central bases of the site. If restriction enzymes do share a common ancestor, as has been proposed, their catalytic regions have been very strongly conserved, while their subunit interfaces and DNA sequence recognition regions have undergone remarkable structural variation.
- W.M. Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, NY 11724.
Organizational Affiliation: