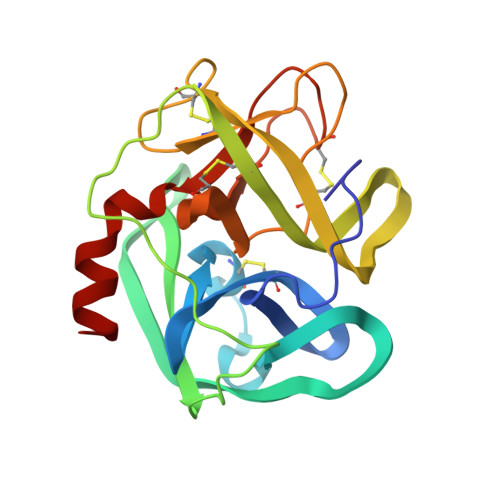
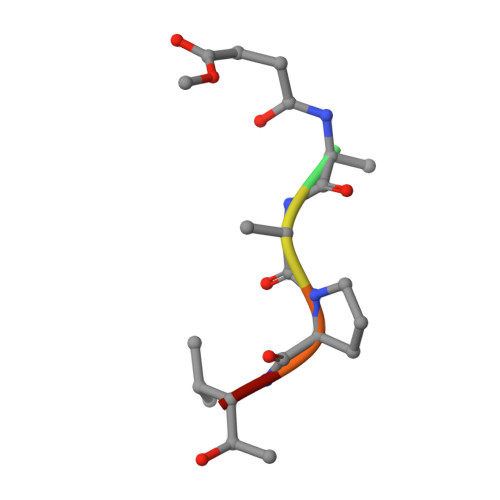
The refined 2.3 A crystal structure of human leukocyte elastase in a complex with a valine chloromethyl ketone inhibitor.
Wei, A.Z., Mayr, I., Bode, W.(1988) FEBS Lett 234: 367-373
- PubMed: 3391280
- DOI: https://doi.org/10.1016/0014-5793(88)80118-2
- Primary Citation of Related Structures:
1PPG - PubMed Abstract:
The stoichiometric complex formed between human leukocyte elastase and a synthetic MeO-Suc-Ala-Ala-Pro-Val chloromethyl ketone inhibitor was co-crystallized and its X-ray structure determined, using Patterson search methods. Its structure has been crystallographically refined to a final R value of 0.145 (8.0 and 2.3 A). The enzyme structure is very similar to that recently observed in a complex formed with the ovomucoid third domain from turkey [(1986) EMBO J. 5,2453-2458]. The rms deviation of all alpha-carbon atoms is 0.32 A. The peptidic inhibitor is bound in a similar overall conformation as the ovomucoid binding segment. Covalent bonds are formed between Val-P1 of the inhibitor and His-57 NE2 and Ser-195 OG of the enzyme. The carbonyl carbon is tetrahedrally deformed to a hemiketal. The valine side chain is arranged in the S1 pocket in the g-conformation.
- Max-Planck-Institut für Biochemie, Martinsried, FRG.
Organizational Affiliation: