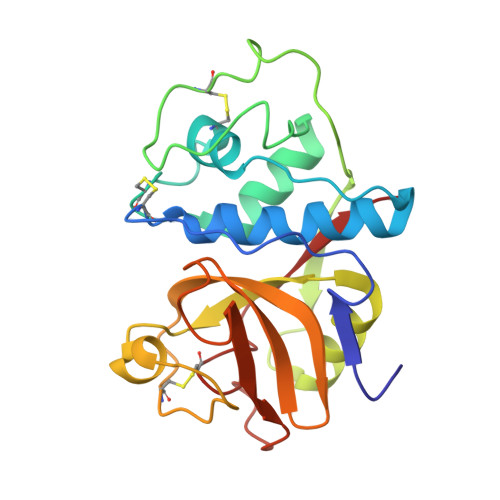
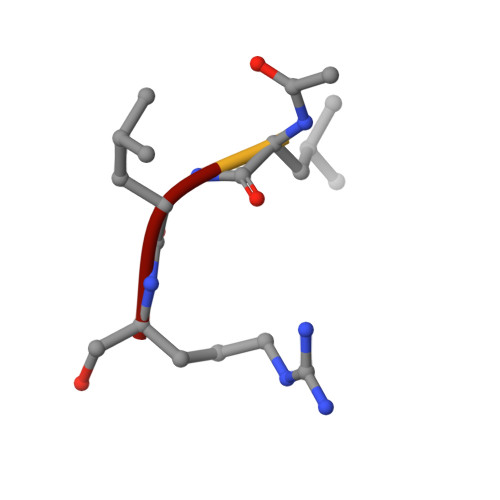
X-ray crystallographic structure of a papain-leupeptin complex.
Schroder, E., Phillips, C., Garman, E., Harlos, K., Crawford, C.(1993) FEBS Lett 315: 38-42
- PubMed: 8416808
- DOI: https://doi.org/10.1016/0014-5793(93)81128-m
- Primary Citation of Related Structures:
1POP - PubMed Abstract:
The three-dimensional structure of the papain-leupeptin complex has been determined by X-ray crystallography to a resolution of 2.1 A (overall R-factor = 19.8%). The structure indicates that: (i) leupeptin contacts the S subsites of the papain active site and not the S' subsites; (ii) the 'carbonyl' carbon atom of the inhibitor is covalently bound by the Cys-25 sulphur atom of papain and is tetrahedrally coordinated; (iii) the 'carbonyl' oxygen atom of the inhibitor faces the oxyanion hole and makes hydrogen bond contacts with Gln-19 and Cys-25.
- University of Oxford, Laboratory of Molecular Biophysics, UK.
Organizational Affiliation: