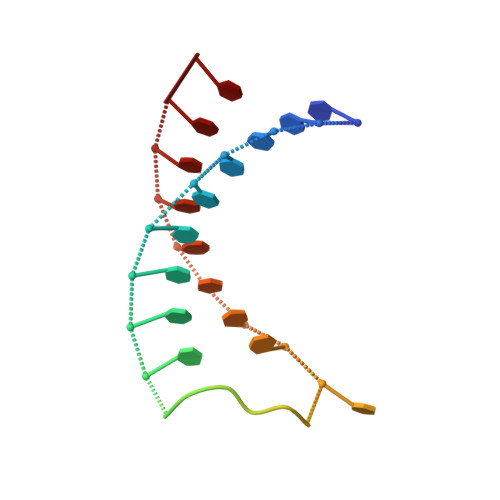
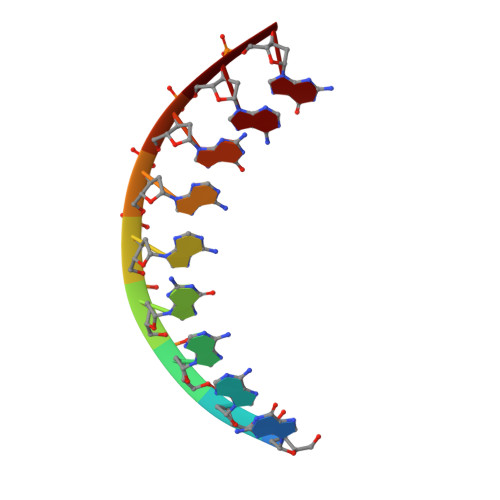
A Nucleic Acid Triple Helix Formed by a Peptide Nucleic Acid-DNA Complex
Betts, L., Josey, J.A., Veal, J.M., Jordan, S.R.(1995) Science 270: 1838-1841
- PubMed: 8525381
- DOI: https://doi.org/10.1126/science.270.5243.1838
- Primary Citation of Related Structures:
1PNN - PubMed Abstract:
The crystal structure of a nucleic acid triplex reveals a helix, designated P-form, that differs from previously reported nucleic acid structures. The triplex consists of one polypurine DNA strand complexed to a polypyrimidine hairpin peptide nucleic acid (PNA) and was successfully designed to promote Watson-Crick and Hoogsteen base pairing. The P-form helix is underwound, with a base tilt similar to B-form DNA. The bases are displaced from the helix axis even more than in A-form DNA. Hydrogen bonds between the DNA backbone and the Hoogsteen PNA backbone explain the observation that polypyrimidine PNA sequences form highly stable 2:1 PNA-DNA complexes. This structure expands the number of known stable helical forms that nucleic acids can adopt.
- Glaxo Wellcome, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: