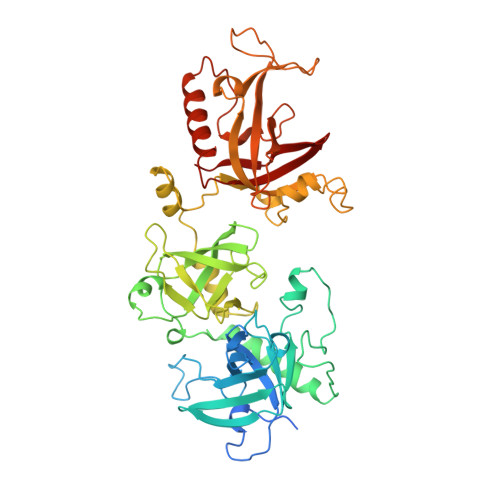
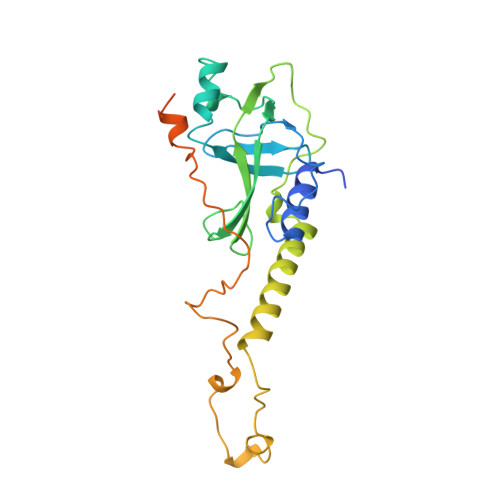
Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA.
Horvath, M.P., Schweiker, V.L., Bevilacqua, J.M., Ruggles, J.A., Schultz, S.C.(1998) Cell 95: 963-974
- PubMed: 9875850
- DOI: https://doi.org/10.1016/s0092-8674(00)81720-1
- Primary Citation of Related Structures:
1OTC - PubMed Abstract:
Telomeres are specialized protein-DNA complexes that compose the ends of eukaryotic chromosomes. Telomeres protect chromosome termini from degradation and recombination and act together with telomerase to ensure complete genome replication. We have determined the crystal structure of the two-subunit Oxytricha nova telomere end binding protein (OnTEBP) complexed with single strand telomeric DNA at 2.8 A resolution. The structure reveals four oligonucleotide/oligosaccharide-binding folds, three of which form a deep cleft that binds the ssDNA, and a fourth that forms an unusual protein-protein interaction between the alpha and beta subunits. This structure provides a molecular description of how the two subunits of OnTEBP recognize and bind ssDNA to form a sequence-specific, telomeric nucleoprotein complex that caps the very 3' ends of chromosomes.
- Department of Chemistry and Biochemistry, University of Colorado, Boulder 80309-0215, USA.
Organizational Affiliation: