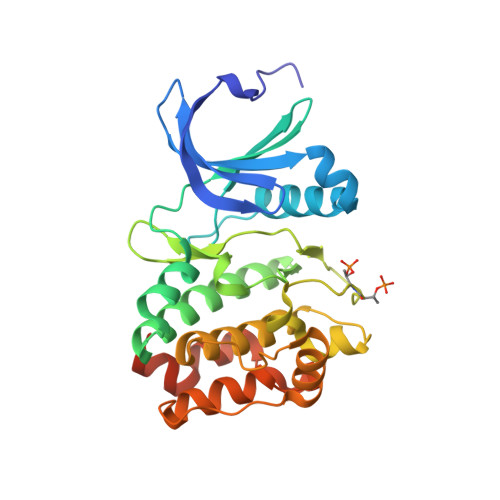
Structural Basis of Aurora-A Activation by Tpx2 at the Mitotic Spindle
Bayliss, R., Sardon, T., Vernos, I., Conti, E.(2003) Mol Cell 12: 851
- PubMed: 14580337
- DOI: https://doi.org/10.1016/s1097-2765(03)00392-7
- Primary Citation of Related Structures:
1OL5, 1OL6, 1OL7 - PubMed Abstract:
Aurora-A is an oncogenic kinase essential for mitotic spindle assembly. It is activated by phosphorylation and by the microtubule-associated protein TPX2, which also localizes the kinase to spindle microtubules. We have uncovered the molecular mechanism of Aurora-A activation by determining crystal structures of its phosphorylated form both with and without a 43 residue long domain of TPX2 that we identified as fully functional for kinase activation and protection from dephosphorylation. In the absence of TPX2, the Aurora-A activation segment is in an inactive conformation, with the crucial phosphothreonine exposed and accessible for deactivation. Binding of TPX2 triggers no global conformational changes in the kinase but pulls on the activation segment, swinging the phosphothreonine into a buried position and locking the active conformation. The recognition between Aurora-A and TPX2 resembles that between the cAPK catalytic core and its flanking regions, suggesting this molecular mechanism may be a recurring theme in kinase regulation.
- Structural and Computational Biology Programme, European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117, Heidelberg, Germany .
Organizational Affiliation: