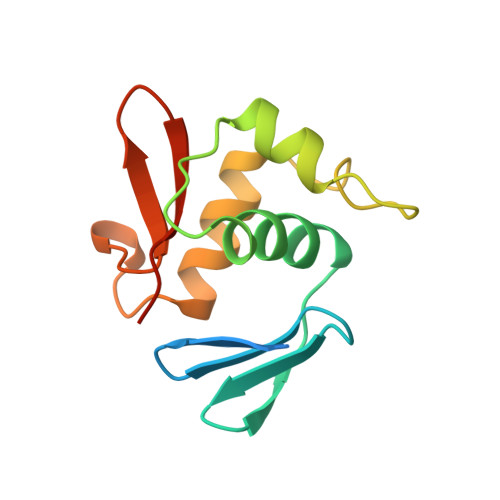
Escherichia coli positive regulator OmpR has a large loop structure at the putative RNA polymerase interaction site.
Kondo, H., Nakagawa, A., Nishihira, J., Nishimura, Y., Mizuno, T., Tanaka, I.(1997) Nat Struct Biol 4: 28-31
- PubMed: 8989318
- DOI: https://doi.org/10.1038/nsb0197-28
- Primary Citation of Related Structures:
1ODD - PubMed Abstract:
The C-terminal DNA-binding domain of OmpR, a positive regulator involved in osmoregulation expression of the ompF and ompC genes in Escherichia coli, has a helix-turn-helix variant motif. The 'turn' region, consisting of 11 residues, forms an RNA polymerase contact site.