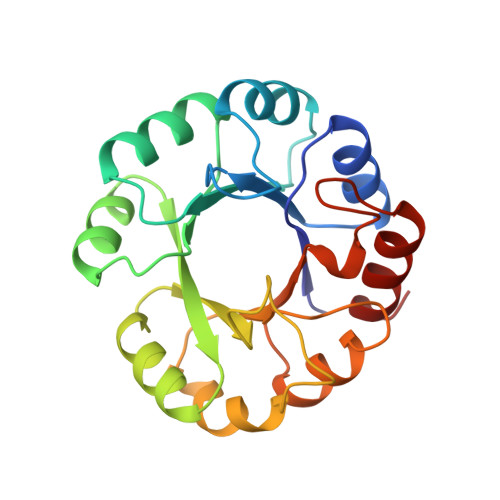
Crystal structure at 2.0 A resolution of phosphoribosyl anthranilate isomerase from the hyperthermophile Thermotoga maritima: possible determinants of protein stability.
Hennig, M., Sterner, R., Kirschner, K., Jansonius, J.N.(1997) Biochemistry 36: 6009-6016
- PubMed: 9166771
- DOI: https://doi.org/10.1021/bi962718q
- Primary Citation of Related Structures:
1NSJ - PubMed Abstract:
The structural basis of thermostability of proteins is of great scientific and biotechnological interest. Differences in the X-ray structues of orthologous proteins from hyperthermophilic and mesophilic organisms can indicate crucial stabilizing interactions. To this end the crystal structure of dimeric phosphoribosyl anthranilate isomerase from the hyperthermophile Thermotoga maritima (tPRAI) was determined using phases derived from the isomorphous replacement method and was refined at 2.0 A resolution. The comparison to the known 2.0 A structure of PRAI from Escherichia coli (ePRAI) shows that tPRAI has the complete TIM- or (beta alpha)8-barrel fold, whereas helix alpha5 in ePRAI is replaced by a loop. The subunits of tPRAI associate via the N-terminal faces of their central beta-barrels. Two long, symmetry-related loops that protrude reciprocally into cavities of the other subunit provide for multiple hydrophobic interactions. Moreover, the side chains of the N-terminal methionines and the C-terminal leucines of both subunits are immobilized in a hydrophobic cluster, and the number of salt bridges is increased in tPRAI. These features appear to be mainly responsible for the high thermostability of tPRAI. In contrast to other hyperthermostable enzymes, tPRAI at 25 degrees C is catalytically more efficient than ePRAI, mainly due to its small K(M) value for the substrate [Sterner, R., Kleemann, G. R., Szadkowski, H., Lustig, A., Hennig, M., & Kirschner, K. (1996) Protein Sci. 5, 2000-2008]. The increased number of hydrogen bonds between the phosphate ion and tPRAI compared to ePRAI could be responsible for this effect.
- Department of Structural Biology, Biozentrum, University of Basel, Switzerland. Michael.Hennig@roche.com
Organizational Affiliation: