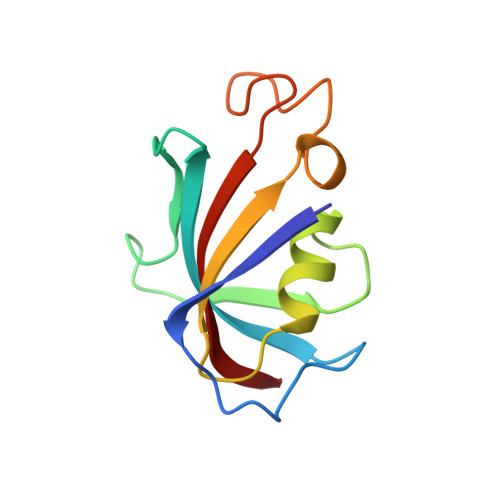
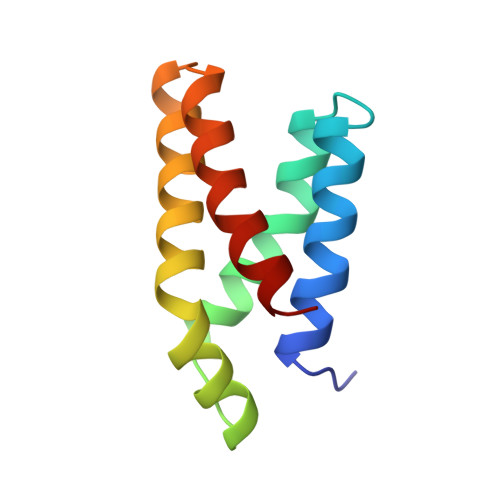
Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Liang, J., Choi, J., Clardy, J.(1999) Acta Crystallogr D Biol Crystallogr 55: 736-744
- PubMed: 10089303
- DOI: https://doi.org/10.1107/s0907444998014747
- Primary Citation of Related Structures:
1NSG, 2FAP, 3FAP, 4FAP - PubMed Abstract:
The structure of the FKBP12-rapamycin-FRB ternary complex has now been refined at 2.2 A resolution. The cell-cycle arrest agent rapamycin binds FK506-binding protein (FKBP12) and the FKBP12-rapamycin binding (FRB) domain of FKBP12-rapamycin associated protein (FRAP) simultaneously, and the inhibition of FRAP is responsible for rapamycin's biological activity. The conformation of rapamycin in the ternary complex is very similar to that observed in the FKBP12-rapamycin binary complex, with an r.m.s. difference of only 0.30 A. However, a slight (9 degrees ) rotation repositions the FRB-binding face of rapamycin in the ternary complex. There are extensive rapamycin-protein interactions and relatively few interactions between the two protein partners FKBP12 and FRB, these interactions mainly involving residues in the 40s and 80s loops of FKBP12 and alpha1 and alpha4 of FRB. The high-resolution refinement has revealed the crucial role of several buried waters in the formation of the ternary complex.
- Department of Chemistry, Cornell University, Ithaca, New York 14853-1301, USA.
Organizational Affiliation: