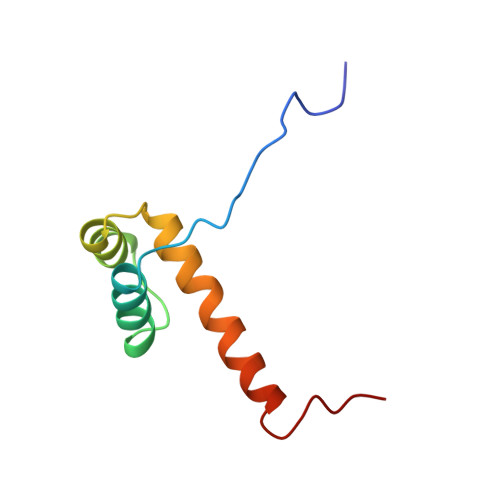
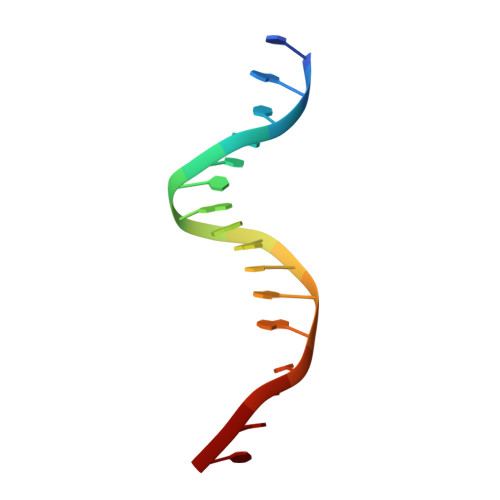
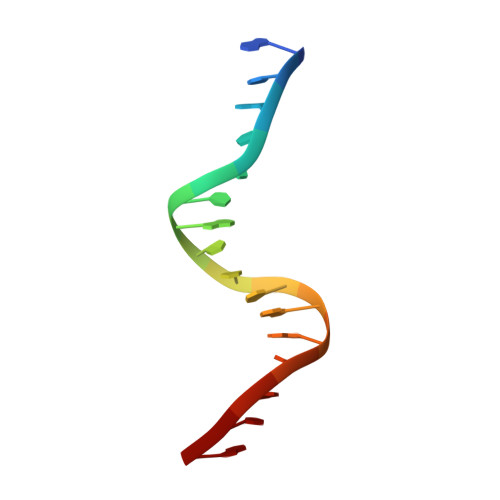
Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Gruschus, J.M., Tsao, D.H., Wang, L.H., Nirenberg, M., Ferretti, J.A.(1997) Biochemistry 36: 5372-5380
- PubMed: 9154919
- DOI: https://doi.org/10.1021/bi9620060
- Primary Citation of Related Structures:
1NK2, 1NK3 - PubMed Abstract:
The interactions responsible for the nucleotide sequence-specific binding of the vnd/NK-2 homeodomain of Drosophila melanogaster to its consensus DNA binding site have been identified. A three-dimensional structure of the vnd/NK-2 homeodomain-DNA complex is presented, with emphasis on the structure of regions of observed protein-DNA contacts. This structure is based on protein-DNA distance restraints derived from NMR data, along with homology modeling, solvated molecular dynamics, and results from methylation and ethylation interference experiments. Helix III of the homeodomain binds in the major groove of the DNA and the N-terminal arm binds in the minor groove, in analogy with other homeodomain-DNA complexes whose structures have been reported. The vnd/NK-2 homeodomain recognizes the unusual DNA consensus sequence 5'-CAAGTG-3'. The roles in sequence specificity and strength of binding of individual amino acid residues that make contact with the DNA are described. We show, based primarily on the observed protein-DNA contacts, that the interaction of Y54 with the DNA is the major determinant of this uncommon nucleotide binding specificity in the vnd/NK-2 homeodomain-DNA complex.
- Laboratory of Biophysical Chemistry, National Heart, Lung, and Blood Institute, Bethesda, Maryland 20892-0380, USA.
Organizational Affiliation: