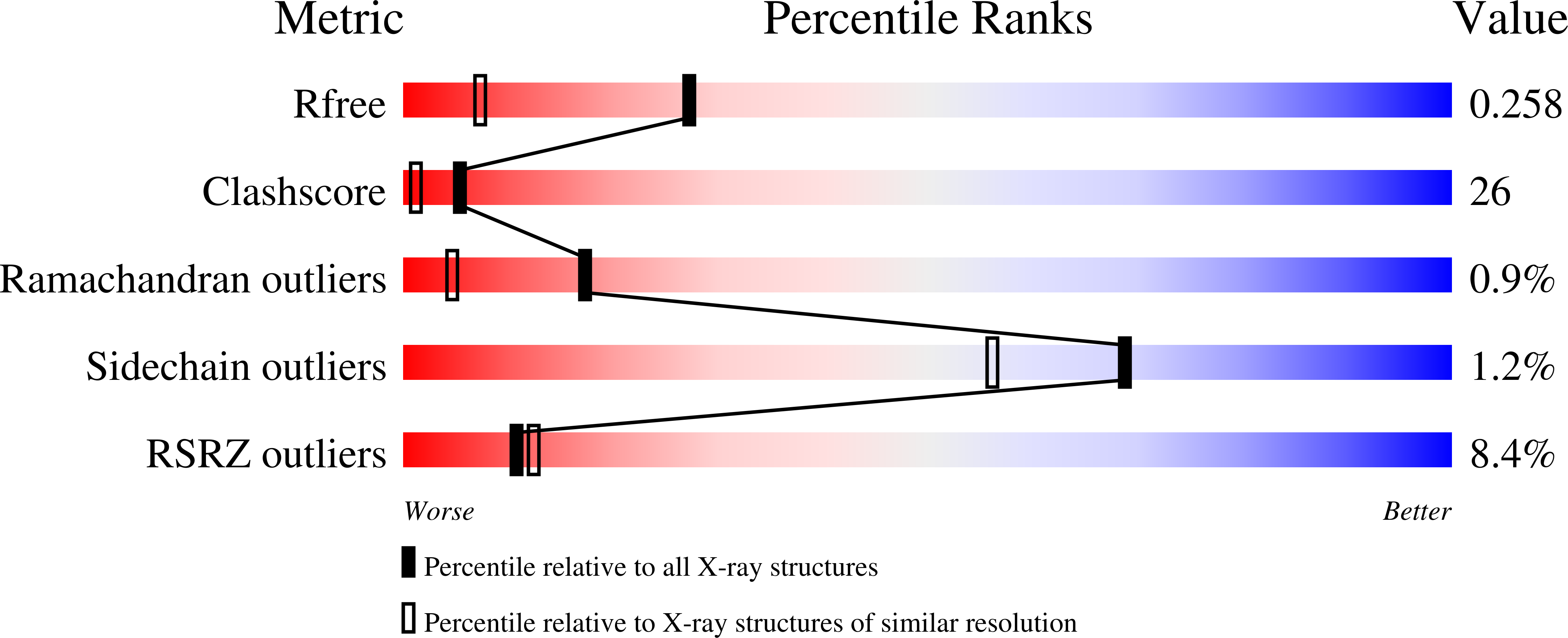
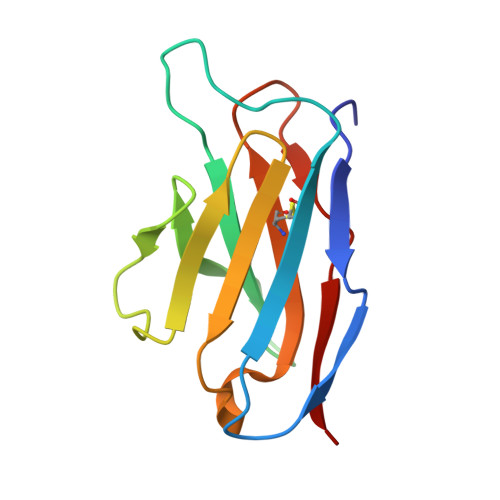
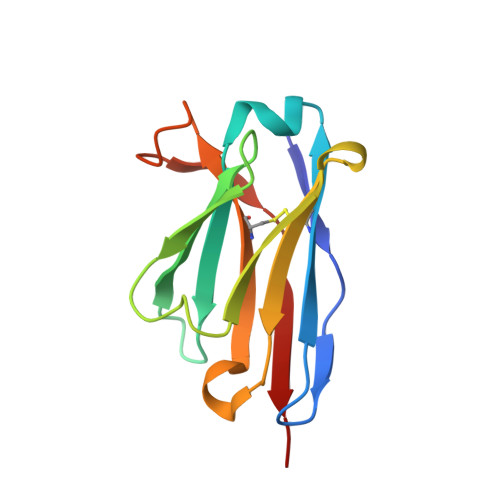
Structure of a single-chain Fv fragment of an antibody that inhibits the HIV-1 and HIV-2 proteases.
Lescar, J., Brynda, J., Fabry, M., Horejsi, M., Rezacova, P., Sedlacek, J., Bentley, G.A.(2003) Acta Crystallogr D Biol Crystallogr 59: 955-957
- PubMed: 12777823
- DOI: https://doi.org/10.1107/s0907444903003597
- Primary Citation of Related Structures:
1N4X - PubMed Abstract:
The monoclonal antibody 1696, which was raised against the HIV-1 protease, inhibits the catalytic activity of the enzyme from both the HIV-1 and HIV-2 strains. The antibody cross-reacts with peptides containing the N-terminus of the enzyme, which is highly conserved between these strains. The crystal structure of a single-chain Fv fragment of 1696 (scFv-1696) in the non-complexed form, solved at 1.7 A resolution, is compared with the previously reported non-complexed Fab-1696 and antigen-bound scFv-1696 structures. Large conformational changes in the third hypervariable region of the heavy chain and differences in relative orientation of the variable domains are observed between the different structures.
- European Synchrotron Radiation Facility, BP 220, F-38043 Grenoble, France.
Organizational Affiliation: