A Y-family polymerase complexed with abasic lesions: catching DNA with a loaded nucleoside triphosphate
Ling, H., Boudsocq, F., Woodgate, R., Yang, W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA polymerase IV | C [auth A] | 352 | Saccharolobus solfataricus P2 | Mutation(s): 0 Gene Names: dpo4 EC: 2.7.7.7 | 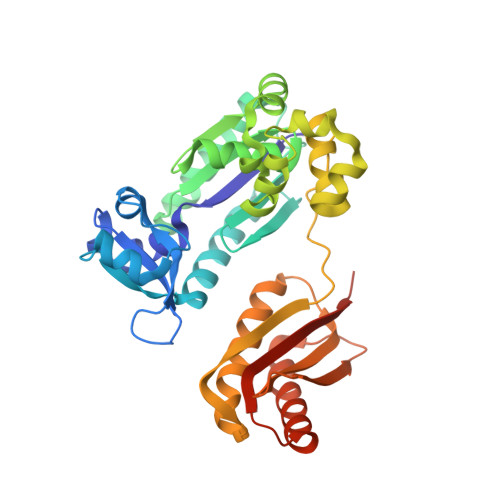 |
UniProt | |||||
Find proteins for Q97W02 (Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2)) Explore Q97W02 Go to UniProtKB: Q97W02 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q97W02 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3' | A [auth B] | 13 | N/A | 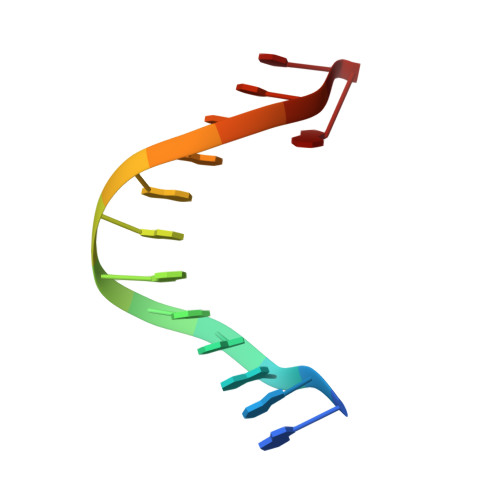 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-D(*CP*AP*(3DR)P*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3' | B [auth C] | 16 | N/A | 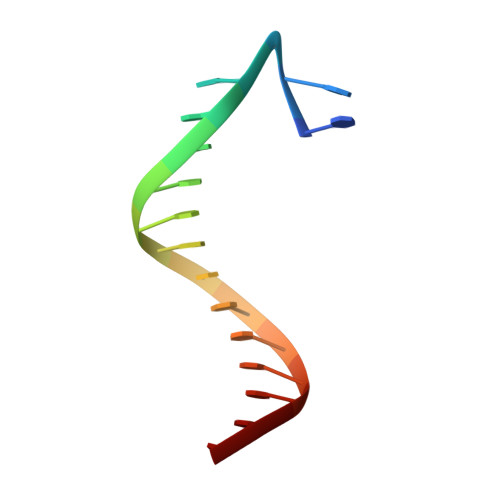 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | E [auth B] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| CA Query on CA | D [auth B], F [auth A], G [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 75.546 | α = 90 |
| b = 88.458 | β = 90 |
| c = 92.389 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |