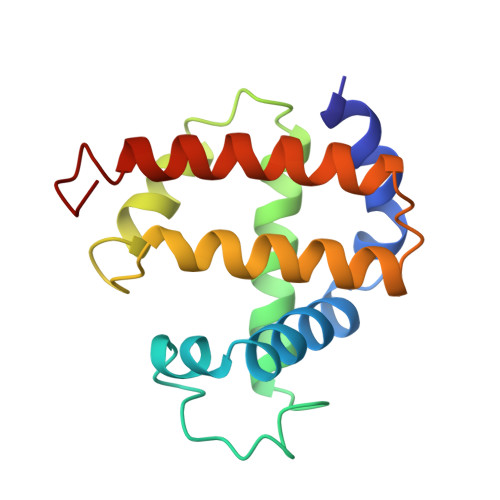
1.70 A resolution structure of myoglobin from yellowfin tuna. An example of a myoglobin lacking the D helix.
Birnbaum, G.I., Evans, S.V., Przybylska, M., Rose, D.R.(1994) Acta Crystallogr D Biol Crystallogr 50: 283-289
- PubMed: 15299440
- DOI: https://doi.org/10.1107/S0907444993014271
- Primary Citation of Related Structures:
1MYT - PubMed Abstract:
The crystal structure of metmyoglobin from yellowfin tuna (Thunnus albacares) has been determined by molecular replacement methods and refined to a conventional R factor of 0.177 for all observed reflections in the range of 6.0-1.70 A resolution. Like other myoglobins for which a high-resolution structure is available, the polypeptide chain is organized into several helices that cooperate to form a hydrophobic pocket into which the heme prosthetic group is non-covalently bound; however, the D helix observed in other myoglobins is absent in myoglobin from yellowfin tuna and has been replaced with a random coil. As well, the A helix has a pronounced kink due to the presence of Pro16. The differences in structure between this and sperm whale myoglobin can be correlated with their reported dioxygen affinity and dissociation. The structure is in agreement with reported fluorescence data which show an increased Trp14.heme distance in yellowfin tuna compared to sperm whale myoglobin.
- Institute for Biological Sciences, National Research Council of Canada, Ottawa.
Organizational Affiliation: