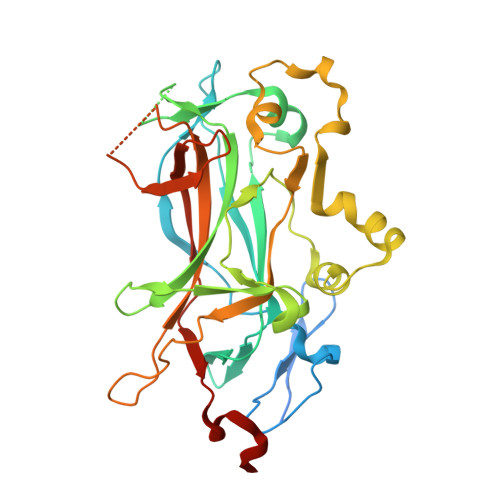
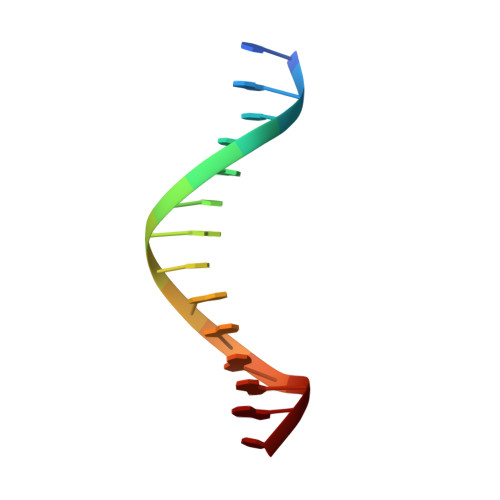
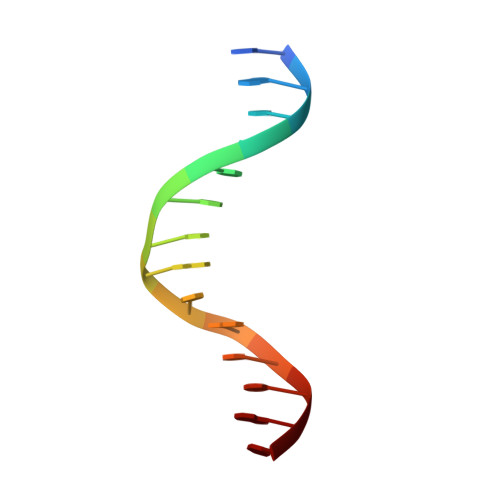
Structure of the sporulation-specific transcription factor Ndt80 bound to DNA
Lamoureux, J.S., Stuart, D., Tsang, R., Wu, C., Glover, J.N.(2002) EMBO J 21: 5721-5732
- PubMed: 12411490
- DOI: https://doi.org/10.1093/emboj/cdf572
- Primary Citation of Related Structures:
1MN4, 1MNN - PubMed Abstract:
Progression through the middle phase of sporulation in Saccharomyces cerevisiae is promoted by the successful completion of recombination at the end of prophase I. Completion of meiotic recombination allows the activation of the sporulation-specific transcription factor Ndt80, which binds to a specific DNA sequence, the middle sporulation element (MSE), and activates approximately 150 genes to enable progression through meiosis. Here, we isolate the DNA-binding domain of Ndt80 and determine its crystal structure both free and in complex with an MSE-containing DNA. The structure reveals that Ndt80 is a member of the Ig-fold family of transcription factors. The structure of the DNA-bound form, refined at 1.4 A, reveals an unexpected mode of recognition of 5'-pyrimidine- guanine-3' dinucleotide steps by arginine residues that simultaneously recognize the 3'-guanine base through hydrogen bond interactions and the 5'-pyrimidine through stacking/van der Waals interactions. Analysis of the DNA-binding affinities of MSE mutants demonstrates the central importance of these interactions, and of the AT-rich portion of the MSE. Functional similarities between Ndt80 and the Caenorhabditis elegans p53 homolog suggest an evolutionary link between Ndt80 and the p53 family.
- Department of Biochemistry, University of Alberta, Edmonton, AB, T6G 2H7, Canada.
Organizational Affiliation: