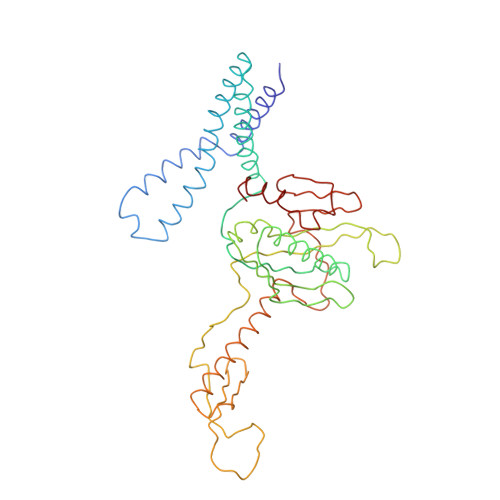
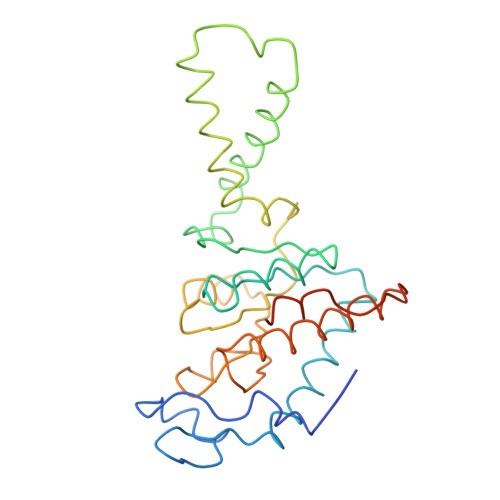
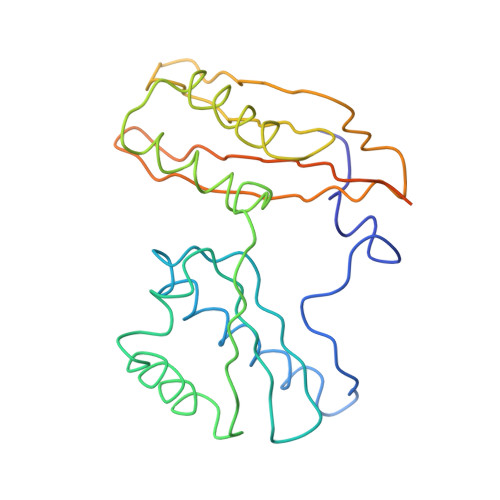
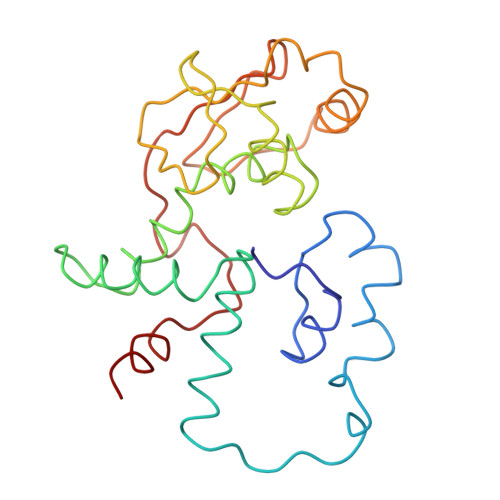
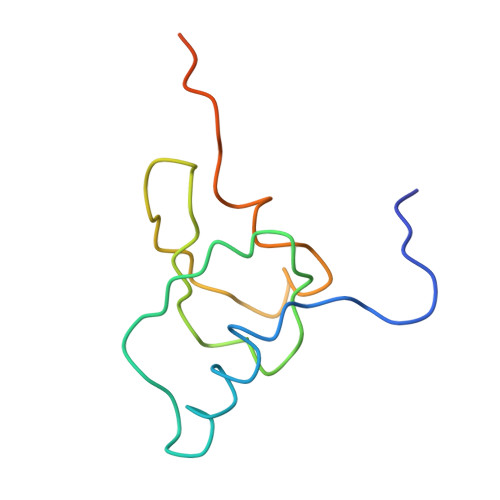
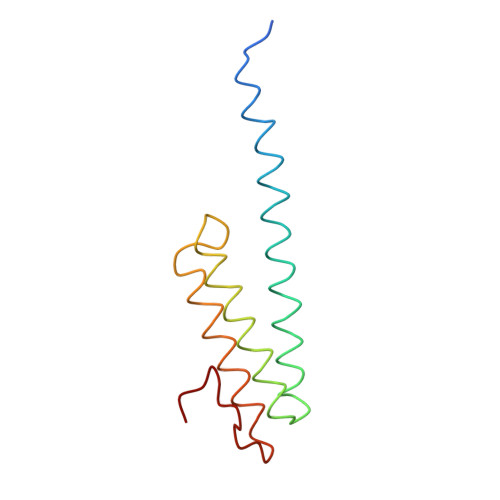
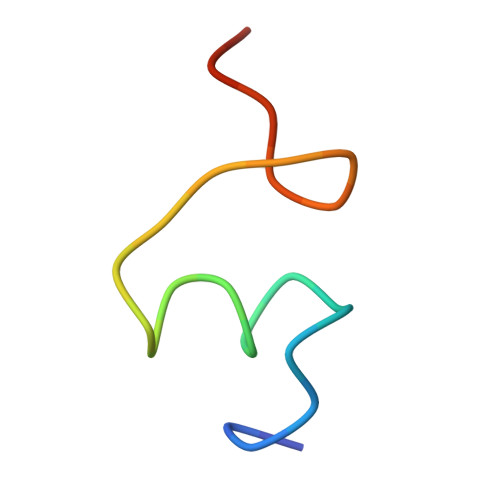
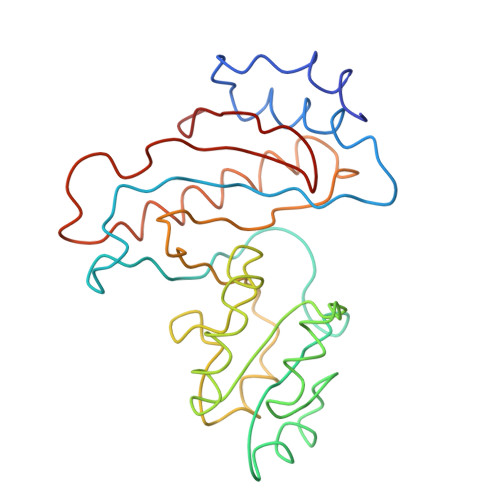
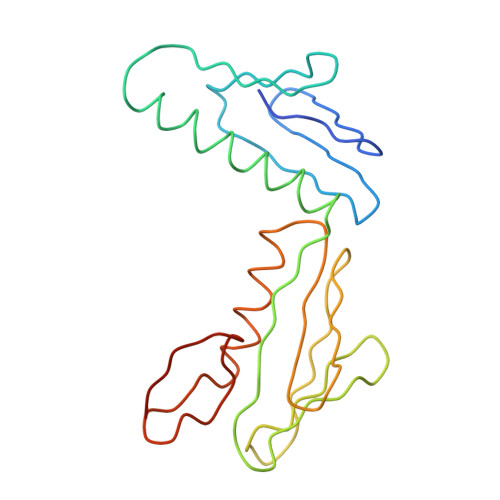
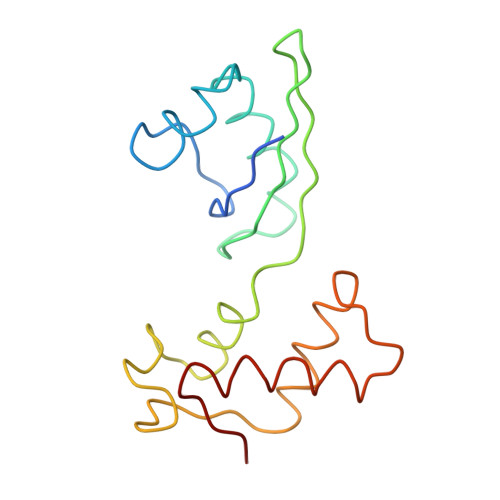
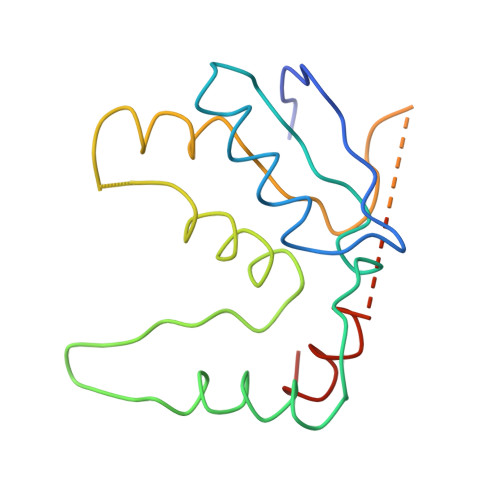
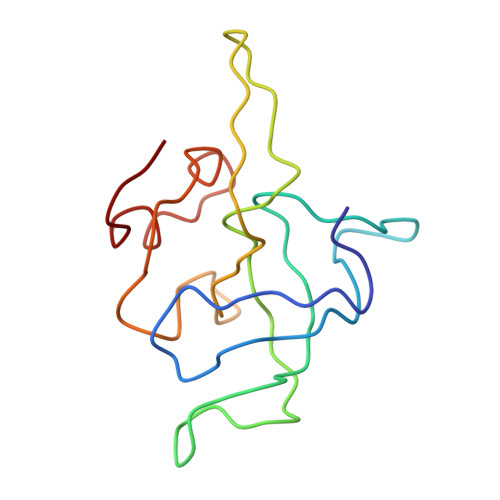
Structure of the Escherichia coli ribosomal termination complex with release factor 2
Klaholz, B.P., Pape, T., Zavialov, A.V., Myasnikov, A.G., Orlova, E.V., Vestergaard, B., Ehrenberg, M., van Heel, M.(2003) Nature 421: 90-94
- PubMed: 12511961
- DOI: https://doi.org/10.1038/nature01225
- Primary Citation of Related Structures:
1ML5 - PubMed Abstract:
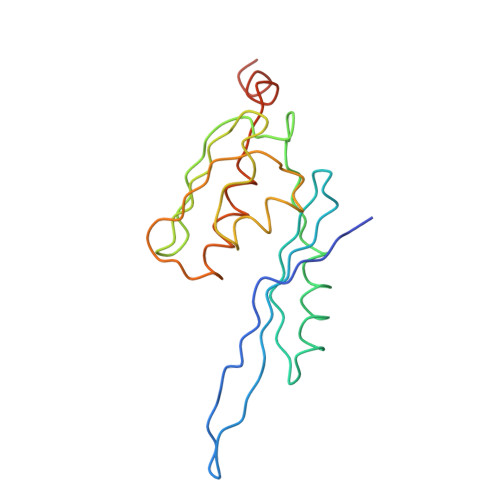
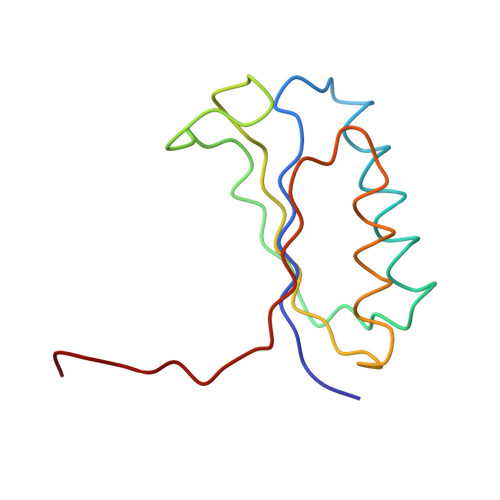
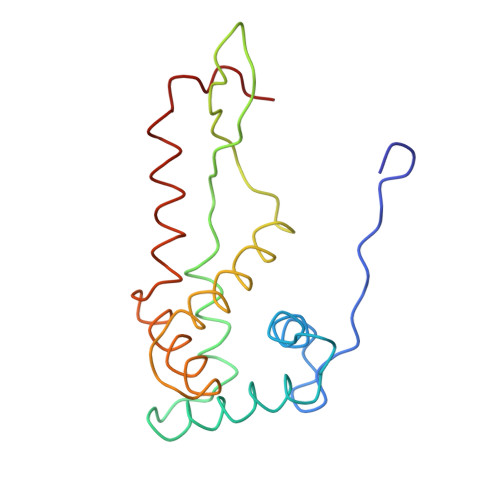
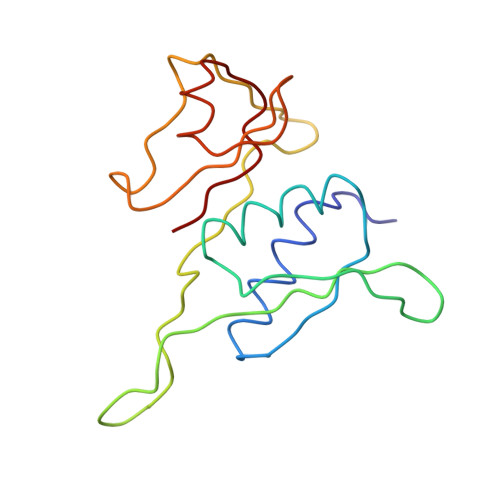
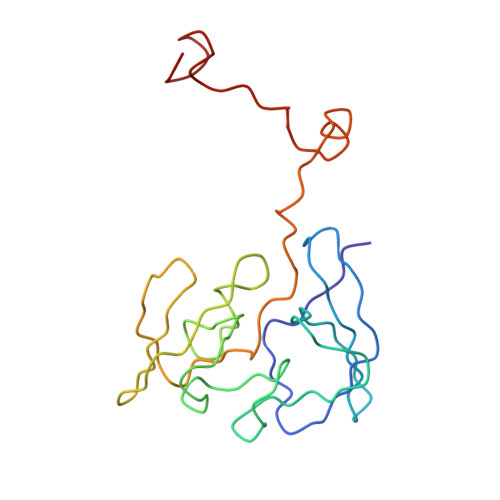
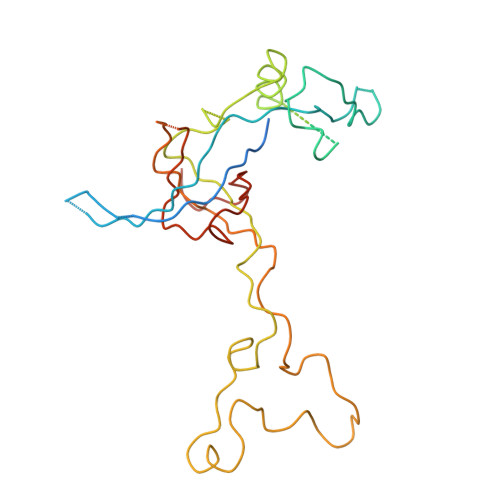
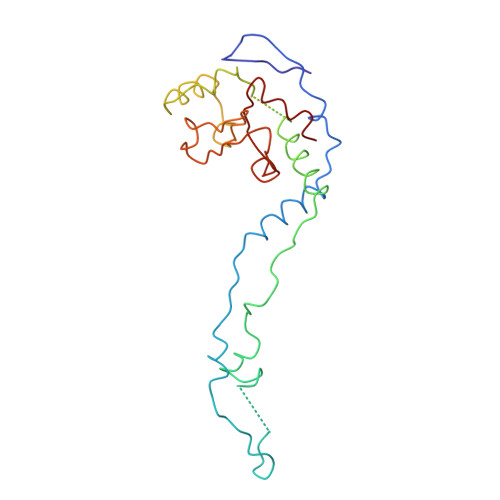
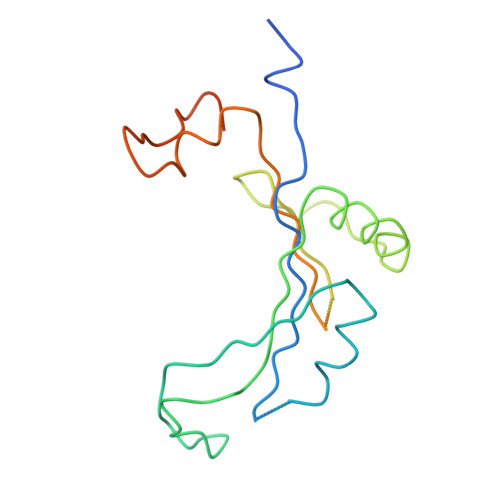
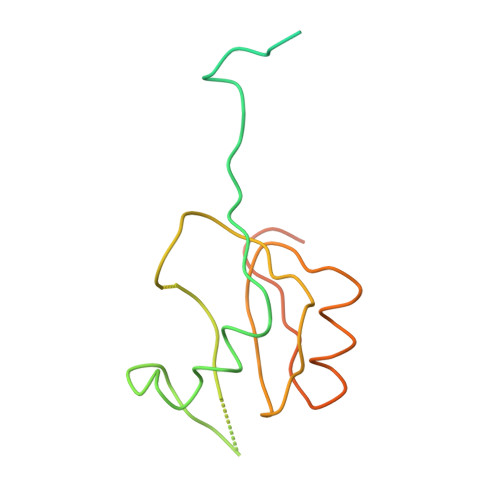
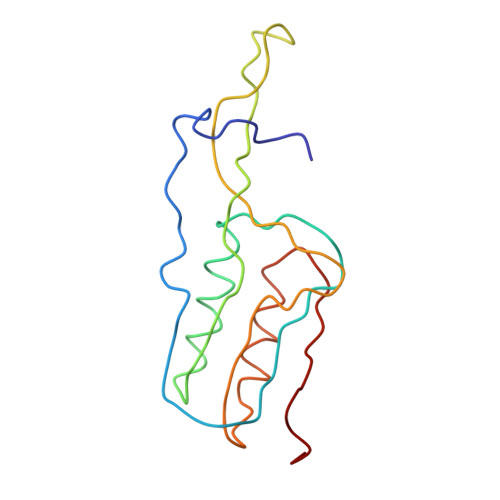
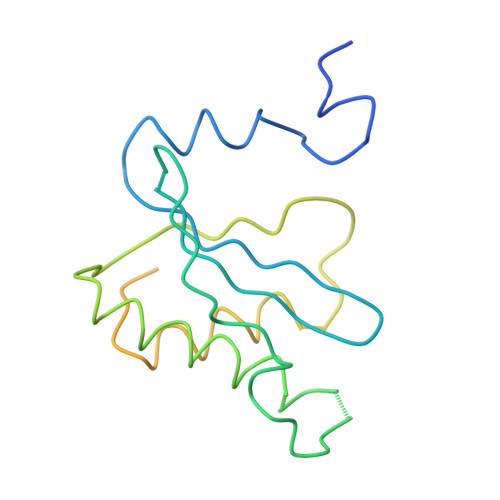
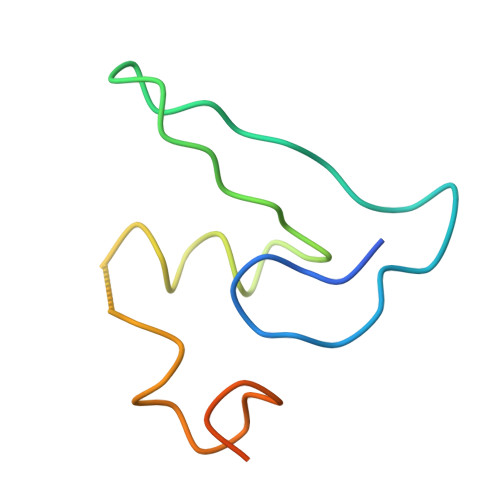
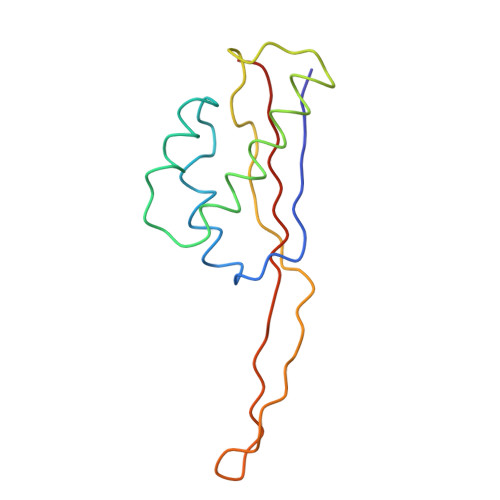
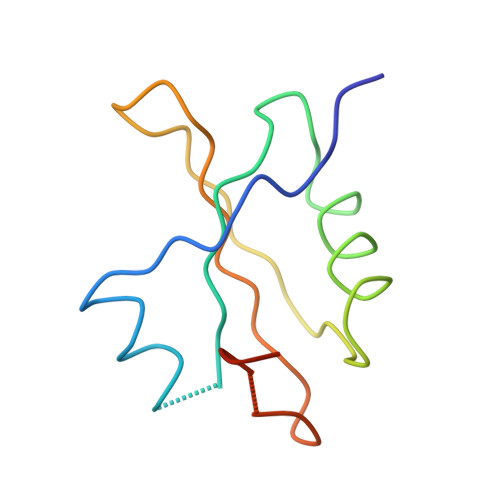
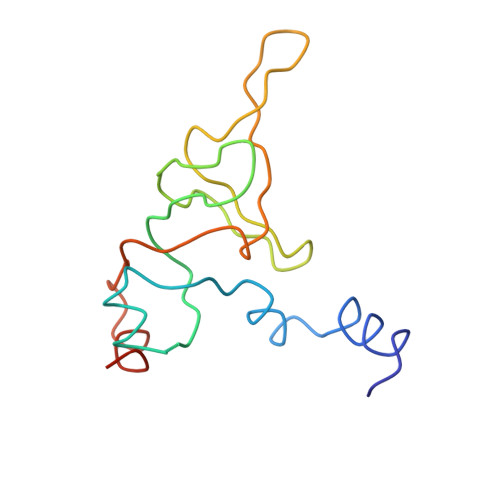
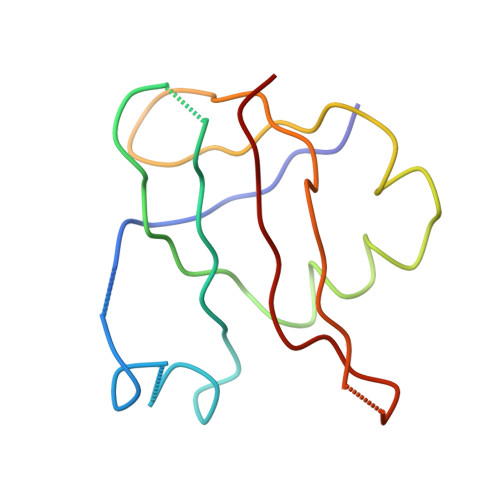
Termination of protein synthesis occurs when the messenger RNA presents a stop codon in the ribosomal aminoacyl (A) site. Class I release factor proteins (RF1 or RF2) are believed to recognize stop codons via tripeptide motifs, leading to release of the completed polypeptide chain from its covalent attachment to transfer RNA in the ribosomal peptidyl (P) site. Class I RFs possess a conserved GGQ amino-acid motif that is thought to be involved directly in protein-transfer-RNA bond hydrolysis. Crystal structures of bacterial and eukaryotic class I RFs have been determined, but the mechanism of stop codon recognition and peptidyl-tRNA hydrolysis remains unclear. Here we present the structure of the Escherichia coli ribosome in a post-termination complex with RF2, obtained by single-particle cryo-electron microscopy (cryo-EM). Fitting the known 70S and RF2 structures into the electron density map reveals that RF2 adopts a different conformation on the ribosome when compared with the crystal structure of the isolated protein. The amino-terminal helical domain of RF2 contacts the factor-binding site of the ribosome, the 'SPF' loop of the protein is situated close to the mRNA, and the GGQ-containing domain of RF2 interacts with the peptidyl-transferase centre (PTC). By connecting the ribosomal decoding centre with the PTC, RF2 functionally mimics a tRNA molecule in the A site. Translational termination in eukaryotes is likely to be based on a similar mechanism.
- Department of Biological Sciences, Imperial College of Science, Technology and Medicine, London SW7 2AY, UK.
Organizational Affiliation: