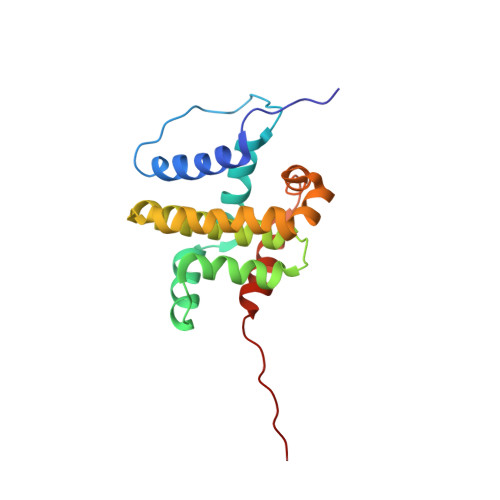
Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix
Denisov, A.Y., Madiraju, M.S., Chen, G., Khadir, A., Beauparlant, P., Attardo, G., Shore, G.C., Gehring, K.(2003) J Biological Chem 278: 21124-21128
- PubMed: 12651847
- DOI: https://doi.org/10.1074/jbc.M301798200
- Primary Citation of Related Structures:
1MK3 - PubMed Abstract:
The structure of human BCL-w, an anti-apoptotic member of the BCL-2 family, was determined by triple-resonance NMR spectroscopy and molecular modeling. Introduction of a single amino acid substitution (P117V) significantly improved the quality of the NMR spectra obtained. The cytosolic domain of BCL-w consists of 8 alpha-helices, which adopt a fold similar to that of BCL-xL, BCL-2, and BAX proteins. Pairwise root meant square deviation values were less than 3 A for backbone atoms of structurally equivalent regions. Interestingly, the C-terminal helix alpha8 of BCL-w folds into the BH3-binding hydrophobic cleft of the protein, in a fashion similar to the C-terminal transmembrane helix of BAX. A peptide corresponding to the BH3 region of the pro-apoptotic protein, BID, could displace helix alpha8 from the BCL-w cleft, resulting in helix unfolding. Deletion of helix alpha8 increased binding affinities of BCL-w for BAK and BID BH3-peptides, indicating that this helix competes for peptide binding to the hydrophobic cleft. These results suggest that although the cytosolic domain of BCL-w exhibits an overall structure similar to that of BCL-xL and BCL-2, the unique organization of its C-terminal helix may modulate BCL-w interactions with pro-apoptotic binding partners.
- Department of Biochemistry and Montreal Joint Center for Structural Biology, McGill University, Montreal, Quebec H3G 1Y6, Canada.
Organizational Affiliation: